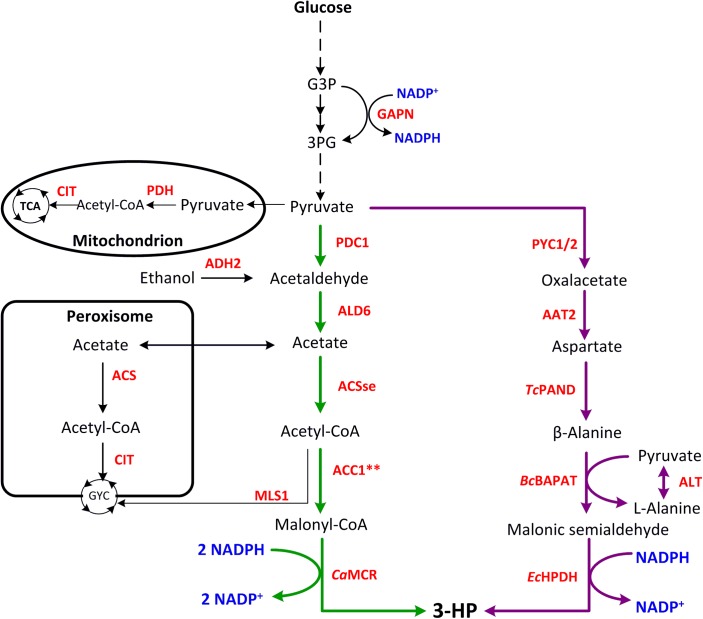
FIGURE 1.
Metabolic pathways constructed in yeast for 3-HP production. Malonyl-CoA reductase pathway is shown in green, and β-alanine pathway is shown in purple. G3P, glyceraldehyde 3-phosphate; 3PG, 3-phosphoglycerate; GYC, glyoxylate cycle; TCA, tricarboxylic acid cycle; AAT2, aspartate aminotransferase 2; ACC1∗∗, Snf1 deregulated acetyl-CoA carboxylase (two site mutations, one is Ser659→Ala, the other is Ser1157→Ala); ACS, acetyl-CoA synthetase; ACSse, acetylation-insensitive acetyl-CoA synthetase from Salmonella enterica; ADH2, alcohol dehydrogenase 2; ALD6, NADP-dependent aldehyde dehydrogenase; ALT, alanine aminotransferase; BcBAPAT, β-alanine-pyruvate aminotransferase from Bacillus cereus; CIT, citrate synthase; EcHPDH, 3-hydroxypropionate dehydrogenase from E. coli; GAPN, NADP-dependent G3P dehydrogenase; CaMCR, malonyl-CoA reductase from Chloroflexus aurantiacus; MLS1, cytosolic malate synthase; PDC1, pyruvate decarboxylase; PDH, pyruvate dehydrogenase complex; PYC1/2, pyruvate carboxylase 1/2; TcPAND, aspartate decarboxylase from Tribolium castaneum.