Abstract
Nutlin-3a is a MDM2 antagonist and preclinical drug that activates p53. Cells with MDM2 gene amplification are especially prone to Nutlin-3a-induced apoptosis, though the basis for this is unclear. Glucose metabolism can inhibit apoptosis in response to Nutlin-3a through mechanisms that are incompletely understood. Glucose metabolism through the pentose phosphate pathway (PPP) produces NADPH that can protect cells from potentially lethal reactive oxygen species (ROS). We compared apoptosis and glucose metabolism in cancer cells with and without MDM2 gene amplification treated with Nutlin-3a. Apoptosis in MDM2-amplified cells was associated with a reduction in glycolysis and the PPP, reduced NADPH, increased ROS, and depletion of the transcription factor SP1, which normally promotes PPP gene expression. In contrast, glycolysis and the PPP were maintained or increased in MDM2 non-amplified cells treated with Nutlin-3a. This was dependent on p53-mediated AKT activation and was associated with maintenance of SP1 and continued expression of PPP genes. Knockdown or inhibition of AKT, SP1, or the PPP sensitized MDM2-non-amplified cells to apoptosis. The data indicate that p53 promotes AKT and SP1-dependent activation of the PPP that protects cells from Nutlin-3a-induced apoptosis. These findings provide insight into how glucose metabolism reduces Nutlin-3a-induced apoptosis, and also provide a mechanism for the heightened sensitivity of MDM2-amplified cells to apoptosis in response to Nutlin-3a.
Keywords: Nutlin, p53, MDM2, SP1, glycolysis, pentose phosphate pathway
Introduction
Wild-type p53 is a tumor suppressor and potent cell growth inhibitor. The activity most associated with tumor suppression by p53 is its ability to bind DNA in a sequence-specific manner and regulate gene transcription. Wild-type p53 is expressed at low levels in most cells due to MDM2, an E3 ubiquitin-ligase enzyme that binds p53 and promotes its degradation (Haupt et al., 1997; Kubbutat et al., 1997). However, the p53 protein is stabilized and its levels increase in response to various stresses, including stresses such as DNA damage and aberrant oncogene signaling that have the potential to promote tumorigenesis (Vousden and Prives, 2009). The stabilization of p53 results from post-translational modifications or induced protein interactions that disrupt p53–MDM2 binding (Sherr, 2006; Kruse and Gu, 2009). The effects of stabilizing p53 and increasing its levels are to halt cell proliferation or induce cell death. These effects are mediated by p53-responsive gene products that arrest the cell cycle (e.g. p21) or induce apoptosis (e.g. PUMA, Noxa, and Bax) (Laptenko and Prives, 2006).
A long standing paradigm was that tumor suppression by p53 resulted solely from its ability to induce cell cycle arrest or apoptosis. However, recent studies indicate p53 can also alter/regulate cancer cell metabolism and that this contributes to tumor suppression (Li et al., 2012; Kruiswijk et al., 2015; Wang and Gu, 2014). Cancer cells display a metabolic reprogramming that includes increased glycolysis and increased dependence on glycolysis for ATP production. Cancer-associated oncoproteins like AKT, c-Myc, and HIF1-α promote glycolysis through transcriptional and non-transcriptional mechanisms (Miller et al., 2012; Ward and Thompson, 2012a). In contrast, p53 can inhibit glycolysis by repressing the expression of multiple glycolytic enzyme genes and/or by increasing expression of TIGAR (Zawacka-Pankau et al., 2011; Kruiswijk et al., 2015). Glucose metabolism can inhibit p53-dependent apoptosis. This is evidenced by findings that 2DG, a non-metabolizable glucose analog, can increase apoptosis in response to p53-activating compounds (Zawacka-Pankau et al., 2011; Duan et al., 2015). One study suggested glucose metabolism inhibits p53-dependent apoptosis by maintaining the expression of ATG genes required for prosurvival autophagy (Duan et al., 2015). Alternatively, glucose metabolism through the pentose phosphate pathway (PPP) could inhibit apoptosis through the generation of NADPH molecules that can combat potentially lethal reactive oxygen species (ROS) (Bensaad et al., 2006). Consistent with this possibility is our report in which we showed a ROS scavenger that can protect p53 wild-type cells from Nutlin-3a-induced apoptosis (Duan et al., 2015).
The role of the transcription factor SP1 in controlling glycolysis and PPP gene expression in p53-activated cells is unclear. For example, SP1 can bind the promoter regions of multiple glycolysis and PPP enzyme genes and promote their expression (Johnson and McLachlan, 1994; Franze et al., 1998; Archer, 2012; Oleaga et al., 2012). However, Zawacka-Pankau et al. (2011) reported that SP1 can also cooperate with p53 to repress the expression of at least some glycolytic enzyme genes. Notably, SP1 is a target for ubiquitination and degradation by MDM2 (Li et al., 2014). This suggests that MDM2 could indirectly regulate glycolysis and/or the PPP by modulating SP1 protein levels. Glucose metabolism/glycolysis is believed to protect cells from p53-mediated apoptosis, though the mechanism for this protection is unclear (Zawacka-Pankau et al., 2011; Duan et al., 2015).
MDM2 gene amplification is often detected in cancers with wild-type p53 (Momand et al., 1998; Oliner et al., 2016). MDM2-amplified cancer cells are especially sensitive to apoptosis by small molecules like Nutlin-3a (Nutlin) that activate p53 by disrupting p53–MDM2 binding (Tovar et al., 2006, 2013). In the current report, we monitored glycolysis and apoptosis in p53 wild-type cancer cell lines treated with Nutlin to activate p53. This included cancer cells with and without MDM2 amplification. Glycolysis and the PPP were inhibited and apoptosis was induced by Nutlin in MDM2-amplified cancer cells. This coincided with high levels of MDM2, depletion of SP1, repression of glycolytic and PPP enzyme genes, reduced NADPH, and increased ROS. In contrast, glycolysis and the PPP were maintained or increased in MDM2-non-amplified cells treated with Nutlin. This was dependent on p53-mediated AKT activation and was associated with maintenance of SP1 and continued expression of glycolysis and PPP genes. Finally, knockdown or inhibition of AKT, SP1, or the PPP sensitized MDM2-non-amplified cells to Nutlin-induced apoptosis. The data suggest that p53 promotes AKT and SP1-dependent activation of the PPP that protects cells from Nutlin-3a-induced apoptosis.
Results
Nutlin-induced p53 promotes or suppresses glycolysis
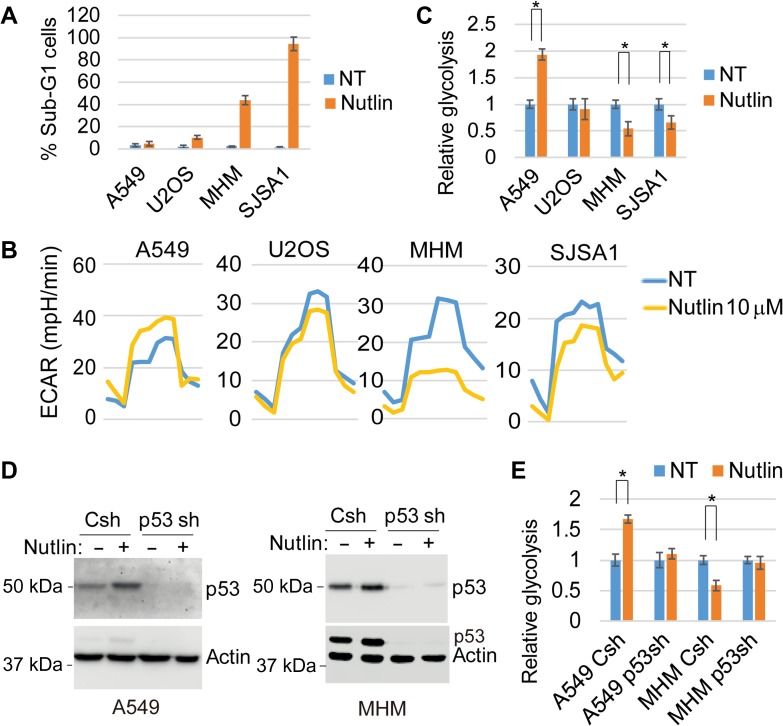
Nutlin-3a (Nutlin) increases wild-type p53 levels by blocking the interaction between p53 and MDM2 (Vassilev et al., 2004). Nutlin-induced p53 can inhibit glycolysis and promote cell cycle arrest and apoptosis (Tovar et al., 2006; Zawacka-Pankau et al., 2011; Duan et al., 2015). Cancer cells with MDM2 amplification are especially prone to Nutlin-induced apoptosis (Tovar et al., 2006, 2013). We compared glycolysis and apoptosis in Nutlin-treated cancer cell lines that express wild-type p53, including cell lines with MDM2 amplification (MHM, SJSA1) and cell lines without MDM2 amplification (U2OS, A549). Apoptosis was determined by the percentage of cells with sub-G1 DNA content. MDM2-amplified MHM and SJSA1 cells are sensitive to apoptosis by Nutlin while MDM2 non-amplified cells U2OS and A549 cells are mostly resistant (Figure 1A). To examine glycolysis, we monitored extracellular acidification rate (ECAR) on a Seahorse Flux analyzer. We found that Nutlin inhibited glycolysis in MHM and SJSA1 cells (Figure 1B and C). Stable knockdown of p53 in MHM cells confirmed the inhibition of glycolysis is p53-dependent (Figure 1D and E). However, Nutlin did not reduce glycolysis in U2OS and, surprisingly, increased glycolysis/ECAR in A549 cells (Figure 1B and C). Increased glycolysis in Nutlin-treated A549 cells was abrogated by p53 knockdown (Figure 1D and E), confirming the increase is p53-dependent.
Figure 1.
Nutlin-induced p53 promotes or suppresses glycolysis. (A) Cells were treated with vehicle or Nutlin (10 μM) for 72 h and analyzed by flow cytometry for sub-G1 cells. Percentage of sub-G1 cells with SD were presented. (B) Cells were treated with vehicle or Nutlin (10 μM) for 24 h and analyzed for glycolysis by Seahorse. (C) Relative glycolysis of Nutlin-treated vs. vehicle-treated was presented with SD indicated. There is significant difference between Nutlin-treated and vehicle-treated A549 cells (P < 0.001), MHM cells (P < 0.05), and SJSA1 cells (P < 0.05), but no significant difference (P > 0.05) in U2OS cells. (D) control and p53 stable knockdown (p53sh) A549 and MHM cells were treated with vehicle or Nutlin (10 μM) for 24 h. Lysates were immunoblotted for the indicated proteins. (E) The cells were also analyzed for glycolysis and relative glycolysis of Nutlin-treated vs. vehicle-treated was presented with SD indicated. There is significant difference between Nutlin-treated and vehicle-treated control A549 cells (P < 0.01) and control MHM cells (P < 0.05), but no significant difference in p53 knockdown A549 and MHM cells (P > 0.05).
AKT promotes glycolysis in response to Nutlin
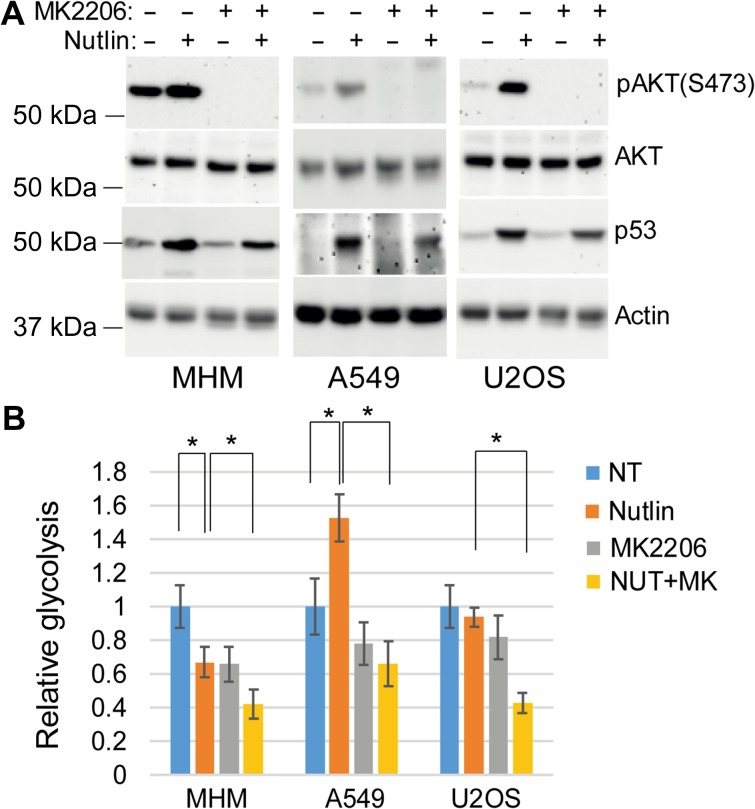
The finding that p53 can increase glycolysis was surprising and we therefore sought the mechanism for this increase. Previous reports indicate that p53 induced by DNA damage or by Nutlin treatment can activate AKT (Singh et al., 2013; Davaadelger et al., 2016). Activated AKT can increase glycolysis through multiple mechanisms including by phosphorylating and activating glycolytic enzymes and by increasing glucose transporter localization to the plasma membrane (Ward and Thompson, 2012b). AKT is activated by phosphorylation at serine 473 (S473) and threonine 308 (T308). AKT was activated by Nutlin treatment, evidenced by increased S473 phosphorylation (Figure 2A). We used the specific AKT inhibitor MK2206 to examine the potential role of AKT in glycolysis in Nutlin-treated cells. MK2206 reduced basal and Nutlin-induced pAKT (S473) levels (Figure 2A) and caused a modest reduction in the basal level of glycolysis in all the cells (Figure 2B), consistent with AKT promoting glycolysis. MK2206 reduced glycolysis in Nutlin-treated MHM and U2OS cells and, most importantly, blocked the increase in glycolysis in Nutlin-treated A549 cells (Figure 2B). This indicates increased glycolysis caused by Nutlin/p53 is AKT-dependent.
Figure 2.
AKT promotes glycolysis in response to Nutlin. (A) Cells were treated with vehicle or Nutlin (10 μM) and/or MK2206 (10 μM) for 24 h. Lysates were immunoblotted for the indicated proteins. (B) The cells were also analyzed for glycolysis and relative glycolysis was presented with SD indicated. There is significant difference between Nutlin-treated and vehicle-treated MHM cells (P < 0.05) and A549 cells (P < 0.01) but no significant difference (P > 0.05) in U2OS cells. There is significant difference between Nutlin-treated and Nutlin plus MK2206-treated MHM (P < 0.05), A549 (P < 0.05), and U2OS (P < 0.05) cells.
Nutlin activates AKT by inhibiting mTORC1 and activating AMPK–TSC2
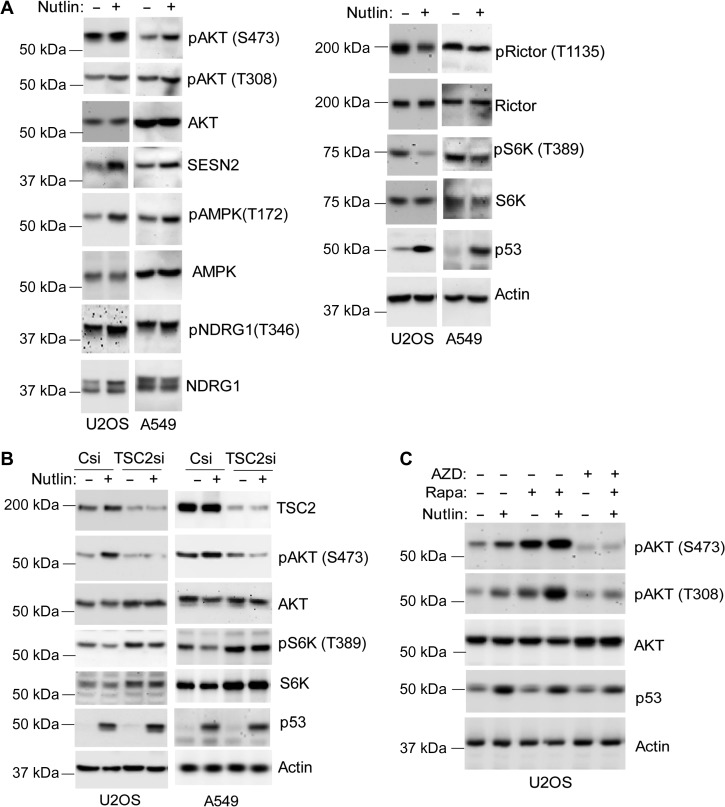
Next, we examined the possible mechanism for AKT activation in Nutlin-treated cells. We considered Nutlin/p53 may activate AKT via the AMPK−TSC2 energy-sensing pathway (Feng and Levine, 2010). p53 induces expression of sestrins 1 and 2 (SESN1/2), which activate AMPK by phosphorylation at T172 (Budanov and Karin, 2008). Active AMPK promotes phosphorylation and activation of TSC2 (Huang and Manning, 2008). TSC2 can bind and activate mTORC2 which can directly phosphorylate AKT at S473 (Huang et al., 2008). TSC2 also inhibits mTORC1 (Huang and Manning, 2009); this inhibition can increase AKT activation by relieving the negative feedback of growth factor signaling mediated by phosphorylated S6K (pS6K) (O’Reilly et al., 2006). pAKT (S473 and T308) induction in Nutlin-treated U2OS and A549 cells was accompanied by increased SESN2 protein and increased phospho-AMPK (T172) levels (Figure 3A). mTORC2 activity increases NDRG1 phosphorylation (Garcia-Martinez and Alessi, 2008). pNDRG1 levels were increased by Nutlin in U2OS and A549 cells (Figure 3A), indicating that mTORC2 was activated. Inhibitory phosphorylation at threonine 1135 (T1135) on Rictor (mTORC2 component) was reduced (Figure 3A), also consistent with mTORC2 activation in Nutlin-treated cells. In contrast, pS6K (T389) levels (indicative of mTORC1 activity) were decreased by Nutlin (Figure 3A). TSC2 is upstream of mTORC1 and mTORC2, and TSC2 siRNA knockdown blocked/reduced pAKT (S473) induction by Nutlin in both U2OS and A549 cells (Figure 3B). This indicates that TSC2 is required for pAKT (S473) induction in Nutlin-treated cells. Direct inhibition of mTORC1 by rapamycin caused a pronounced increase in pAKT (S473) levels in Nutlin-treated cells (Figure 3C), probably by relieving the negative feedback of growth factor signaling that is mediated by mTORC1-pS6K. In contrast, the dual mTORC1/mTORC2 inhibitor AZD8055 (AZD) blocked the Nutlin-induced increase in pAKT(S473) levels while only slightly reducing pAKT (T308), supporting the idea that mTORC2 is required for pAKT (S473) (Figure 3C). Taken together, these results suggest that p53 could increase AKT activation in Nutlin treated through at least two mechanisms: (i) by activating AMPK–TSC2 and increasing mTORC2 activity, and (ii) by inhibiting mTORC1 and relieving feedback inhibition of growth factor signaling mediated by pS6K.
Figure 3.
Nutlin activates AKT by inhibiting mTORC1 and activating AMPK−TSC2. (A) U2OS and A549 cells were treated with Nutlin (10 μM) for 24 h. Lysates were immunoblotted for the indicated proteins. (B) U2OS and A549 cells were transfected with control siRNA or TSC2 siRNA and then treated with vehicle or Nutlin (10 μM) for 24 h. Lysates were immunoblotted for the indicated proteins. (C) U2OS cells were treated with vehicle, Nutlin, or Nutlin plus AZD8055 (100 nM), OSI906 (1 μM), or rapamycin (10 nM) for 24 h. Lysates were immunoblotted for the indicated proteins.
Nutlin decreases glycolytic genes via MDM2-mediated downregulation of SP1
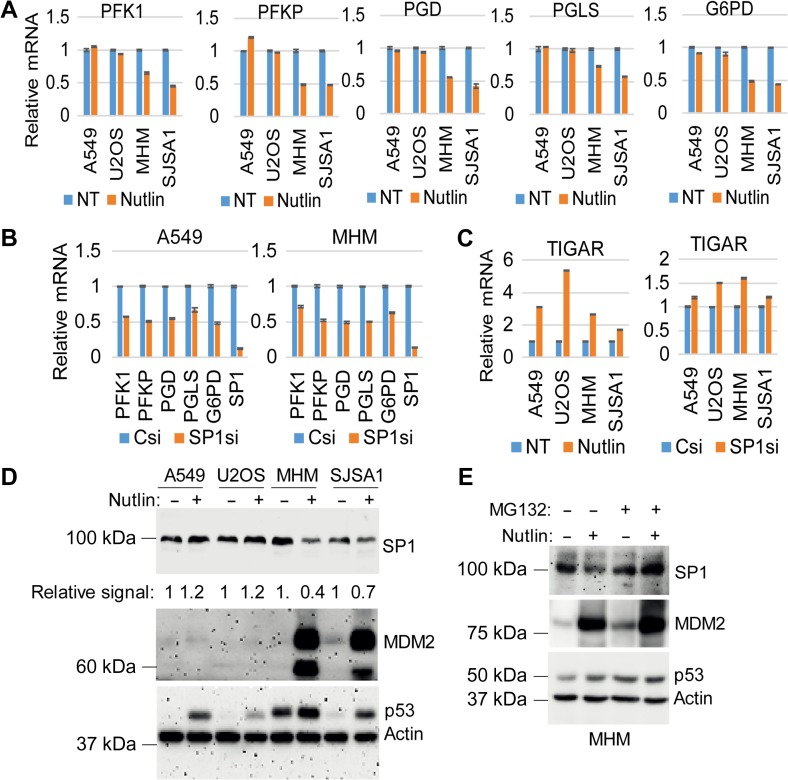
The data in Figure 2 suggest that p53 can increase glycolysis by activating AKT. However, glycolysis was inhibited in Nutlin-treated MHM and SJSA1 cells despite the fact that AKT was activated in these cells. This suggests that other mechanisms must be at play in Nutlin-treated MHM and SJSA1 cells that inhibit glycolysis even when AKT is activated. One of the proposed ways for how p53 can inhibit glycolysis is by reducing the expression of glycolytic enzyme genes (Zawacka-Pankau et al., 2011; Duan et al., 2015). We analyzed the expression of multiple glycolytic genes in Nutlin-treated cells. We found mRNA levels for PFK1, PFKP, PGD, PGLS, and G6PD were down-regulated by Nutlin in MHM and SJSA1 cells but not U2OS and A549 cells (Figure 4A). PFK1/P, PGD, and G6PD were previously shown to be activated by SP1 (Archer; Johnson and McLachlan, 1994; Franze et al., 1998; Oleaga et al., 2012), and SP1 is a target for MDM2-mediated ubiquitination and degradation (Li et al., 2014). SP1 depletion by siRNA reduced the expression of all five genes (Figure 4B), indicating that SP1 is a positive regulator of these genes. p53 can also regulate glycolysis by inducing the expression of TIGAR. TIGAR expression was increased by Nutlin in all of the cell lines and, notably, the increase in TIGAR was most prominent in U2OS cells (Figure 4C). SP1 did not appear to increase TIGAR expression since TIGAR mRNA levels were slightly increased and not decreased in SP1 knockdown cells (Figure 4C). Next we analyzed MDM2 and SP1 protein levels. MDM2 was induced to high levels and SP1 decreased by Nutlin in MHM and SJSA1 cells (Figure 4D). However, MDM2 was only slightly increased and SP1 was slightly increased in U2OS and A549 cells (Figure 4D). Furthermore, proteasome inhibition by MG132 treatment blocked Nutlin-induced decrease in SP1 and increased MDM2 levels in MHM cells (Figure 4E). The data suggest that high levels of MDM2 in MDM2-amplified cells (MHM, SJSA1) lead to depletion/degradation of SP1, and this causes repression of glycolytic enzyme genes.
Figure 4.
Nutlin decreases glycolytic genes via MDM2-mediated downregulation of SP1. (A) Cells were treated with Nutlin (10 μM) for 24 h. mRNA was analyzed by qPCR for the indicated genes. (B) Cells were transfected with control siRNA or SP1 RNA for 48 h. mRNA was analyzed by qPCR for the indicated genes. (C) Cells were treated with vehicle or Nutlin (10 μM) for 24 h, or transfected with control siRNA or SP1 RNA for 48 h. mRNA was analyzed by qPCR for TIGAR. Relative mRNA level is presented as graphs with SD indicated (triplicate). (D) Cells were treated with vehicle or Nutlin (10 μM) for 24 h. Lysates were immunoblotted for the indicated proteins. (E) MHM cells were treated with vehicle or Nutlin (10 μM) for 21 h and then additionally with vehicle or MG132 (10 μM) for 3 h. Lysates were immunoblotted for the indicated proteins.
Nutlin inhibits or promotes PPP
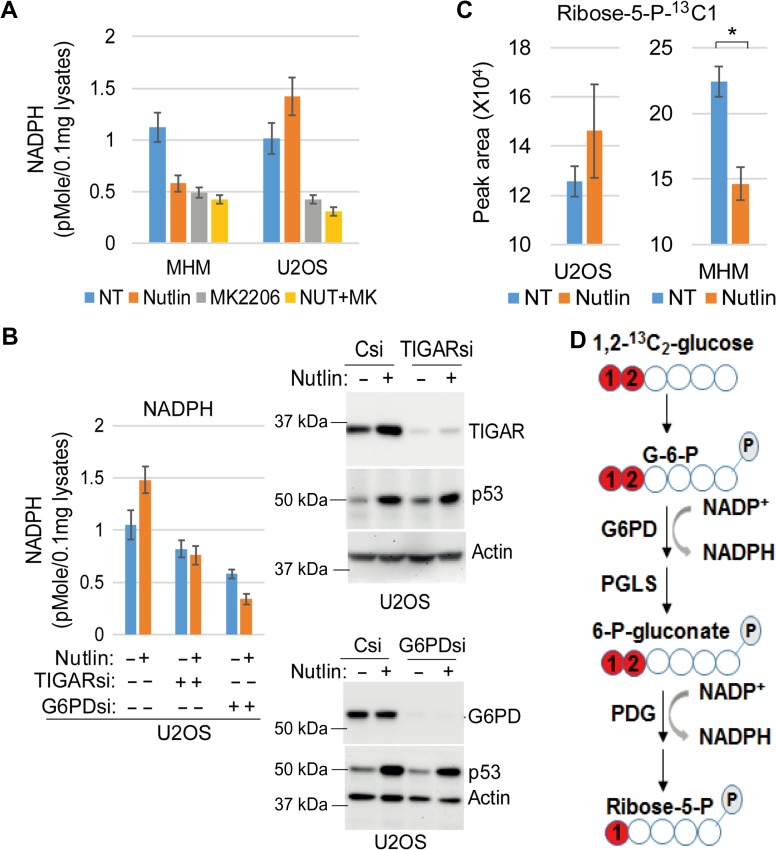
Among the SP1-regulated genes, G6PD, PGD, and PGLS regulate glucose metabolism through the pentose phosphate pathway (PPP). G6PD is the rate-limiting enzyme in the PPP that converts glucose 6-phosphate into 6-phosphogluconolactone while generating one NADPH. PGLS converts 6-phosphogluconolactone into 6-phosphogluconate, and PGD converts 6-phosphogluconate into ribose-5-P while generating one NADPH. NADPH is an important cellular antioxidant. p53-mediated expression of TIGAR is known to promote the PPP and reduce ROS to promote cell survival in p53-activated cells (Bensaad et al., 2006). Therefore, MDM2-mediated downregulation of SP1 and subsequent repression of G6PD, PGD, and PGLS may lead to suppression of PPP and decreasing NADPH in MDM2-amplified cells. To examine this possibility, we measured NADPH in MDM2-amplified MHM cells and MDM2-non-amplified U2OS cells. NADPH was decreased in Nutlin-treated MHM cells (Figure 5A). In contrast, NADPH was increased in Nutlin-treated U2OS cells (Figure 5A). Inhibition of AKT decreased basal NADPH in MHM and U2OS cells and blocked the Nutlin-induced increase in NADPH in U2OS cells (Figure 5A). Knockdown of TIGAR or G6PD lowered basal NADPH levels in U2OS cells and blocked the increase in NADPH levels caused by Nutlin (Figure 5B). This result suggests that Nutlin increases NADPH levels in U2OS cells via the PPP in a manner dependent on AKT, TIGAR, and G6PD. We also analyzed glucose flux by labeling vehicle or Nutlin-treated MHM and U2OS cells with 1,2-13C2-glucose and quantifying levels of the PPP metabolite ribose-5-P-13C1 with LC–MS. The results showed that ribose-5-P-13C1 was slightly increased (P = 0.22) in Nutlin-treated U2OS cells while significantly decreased (P = 0.008) in Nutlin-treated MHM cells (Figure 5C). Ribose-5-P-13C1 is a direct metabolite of 1,2-13C2-glucose (Figure 5D). Together, the results in Figure 5A−C suggest that Nutlin inhibits the PPP and NADPH production in MDM2-amplified cells (e.g. MHM) while promoting the PPP and increasing NADPH production in MDM2-nonamplifed cells (e.g. U2OS).
Figure 5.
Nutlin inhibits or promotes PPP. (A) Cells were treated with vehicle or Nutlin (10 μM) and/or MK2206 (10 μM) for 24 h. Lysates were analyzed for NADPH. Average NADPH level from triplicate was presented with SD indicated. (B) Cells were transfected with control siRNA, TIGAR siRNA, or G6PD siRNA and then treated with vehicle or Nutlin (10 μM) for 24 h. Lysates were analyzed for NADPH. Average NADPH level from triplicate was presented with SD indicated (left panel). Lysates were also immunoblotted for the indicated proteins (right panel). (C) cells were treated with Nutlin for a 12-h period and then fluxed with D-[1,2-13C] glucose for 15 min. Metabolites were analyzed by LC–MS. Ribose-5-P-13C1 levels in vehicle-treated and Nutlin-treated cells were presented with SD indicated. There is no significant difference between vehicle and Nutlin in U2OS cells (P = 0.22). There is significant difference between vehicle and Nutlin in MHM cells (P = 0.008). (D) Metabolism of 1,2-13C2-glucose through PPP to generate NADPH and into ribose-5-P-13C1 is schematically presented.
Depletion of G6PD and SP1 or inhibition of AKT increases ROS and sensitizes U2OS cells to Nutlin-induced apoptosis
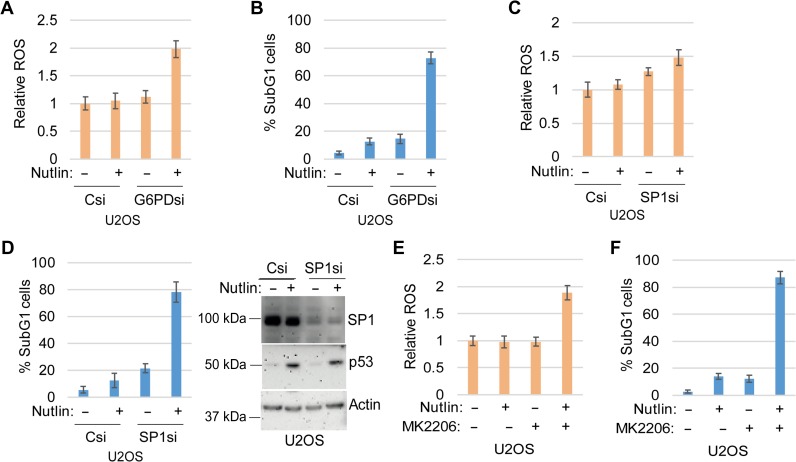
We previously showed that a ROS scavenger can protect cells from Nutlin-induced apoptosis (Duan et al., 2015). NADPH produced in the PPP can counteract ROS. Our data indicate that AKT promotes glucose metabolism in Nutlin-treated cells (Figure 2) and that SP1 promotes PPP gene expression (Figure 4). Based on these data, we speculated that knockdown or inhibition of AKT, SP1, or the PPP would increase ROS and sensitize cells to Nutlin-induced apoptosis. To test this, we knocked down SP1 or G6PD in U2OS cells, or treated the cells with MK2206 (AKT inhibitor), and then measured ROS levels and apoptosis in response to Nutlin. As shown in Figure 6, Nutlin caused little or no increase in ROS in U2OS cells transfected with control siRNA (Csi) or U2OS cells treated with Nutlin alone. However, Nutlin treatment increased ROS when SP1 and G6PD were depleted by siRNA (Figure 6A and C) or AKT was inhibited by MK2206 treatment (Figure 6E). Furthermore, Nutlin treatment caused little apoptosis in control U2OS cells but caused abundant apoptosis when SP1 or G6PD were depleted (Figure 6B and D) or in the presence of MK2206 (Figure 6F). These data support the idea that AKT, SP1, and the PPP prevent ROS accumulation in Nutlin-treated U2OS cells and this protects the cells from apoptosis.
Figure 6.
Depletion of G6PD and SP1 or inhibition of AKT increases ROS and sensitizes U2OS cells to Nutlin-induced apoptosis. (A−D) Cells were transfected with control siRNA or G6PD siRNA (A and B) or SP1 siRNA (C and D) and then treated with vehicle or Nutlin (10 μM). Twenty-four hours later, an equal number of cells were analyzed for ROS. Relative ROS level was presented with SD indicated (A and C). Seventy-two hours later, the cells were analyzed by flow cytometry. Percentage of sub-G1 cells was presented with SD indicated (B and D). (E and F) Cells treated with vehicle or Nutlin (10 μM) and/or MK2206 (10 μM). Twenty-four hours later, an equal number of cells were analyzed for ROS. Relative ROS level was presented with SD indicated (E). Seventy-two hours later, the cells were analyzed by flow cytometry. Percentage of sub-G1 cells was presented with SD indicated (F).
Discussion
Glucose can be metabolized to pyruvate in glycolysis or, alternatively, can be shunted to the pentose phosphate pathway (PPP) (Patra and Hay, 2014). Glucose metabolism in the PPP produces NADPH molecules that can combat potentially lethal reactive oxygen species (ROS) (Patra and Hay, 2014). Previous studies showed that blocking glucose metabolism with the non-metabolizable glucose analog 2-DG could sensitize cells to apoptosis by Nutlin (Duan et al., 2015). These results indicated that glucose metabolism protects cells against Nutlin-induced apoptosis. However, it was unclear from these previous studies if the protective effect of glucose metabolism was due to glycolysis per se (glucose to pyruvate metabolism) or from glucose metabolism in the PPP. In the current study, Nutlin treatment increased the PPP and increased NADPH levels in MDM2-non-amplified cells that are resistant to Nutlin-induced apoptosis. Direct inhibition of the PPP by G6PD knockdown reduced NADPH levels, increased ROS, and sensitized the cells to Nutlin-induced apoptosis. Our previous study showed a ROS scavenger can protect cells from apoptosis by Nutlin (Duan et al., 2015). Together, the findings indicate that glucose metabolism through the PPP can protect cells from Nutlin-induced apoptosis, most likely by increasing NADPH levels to counteract ROS. It remains possible that glucose metabolism through glycolysis (glucose metabolism to pyruvate) also protects against Nutlin-induced apoptosis.
AKT can promote survival by phosphorylating and altering the activity of various pro- and anti-apoptotic factors (Song et al., 2005). AKT can also increase glucose metabolism by promoting translocation of glucose transporters to the plasma membrane and phosphorylating and increasing the activity of certain glycolysis enzymes (Ward and Thompson, 2012a, b). In the current study we found AKT was activated in Nutlin-treated cells. Our data suggest AKT activation could result from mTORC1 inhibition in Nutlin-treated cells and/or activation of AMPK–TSC2–mTORC2 signaling. AKT inhibition reduced glycolysis and reduced NADPH production via the PPP in Nutlin-treated cells. This was associated with increased ROS and sensitization to Nutlin-induced apoptosis. Based on this, we propose that AKT can protect cells against Nutlin-induced apoptosis, at least in part, by increasing glucose metabolism and NADPH production via the PPP. Notably, Nutlin-induced phosphorylation (activation) of AKT to higher levels in U2OS cells than that in A549 cells, while Nutlin-induced glycolysis to higher levels in A549 cells than in U2OS cells. This suggests that AKT activation is not the only factor that determines glycolysis in response to Nutlin. p53 can inhibit glycolysis by inducing expression of TIGAR that shunts glucose metabolism into the PPP pathway (Bensaad et al., 2006). TIGAR gene expression was induced to a higher level in response to Nutlin in U2OS cells than in A549 cells, which may offer some explanation for the glycolysis difference between A549 and U2OS cells.
MDM2-amplified cancer cells are especially sensitive to apoptosis in response to MDM2 antagonists like Nutlin. The basis for this heightened apoptosis sensitivity is not clear. The transcription factor SP1 is a target for MDM2-mediated degradation (Li et al., 2014). SP1 can promote the expression of various glycolysis and PPP enzyme genes through SP1-binding sites in their promoters (Johnson and McLachlan, 1994; Franze et al., 1998; Archer, 2012; Oleaga et al., 2012). Nutlin treatment caused a pronounced increase in MDM2 and depletion of SP1 in MDM2-amplified cancer cells but not MDM2-non-amplified cells. MG132 restored SP1 in Nutlin-treated MHM cells, suggesting MDM2-mediated degradation of SP1 via proteasomes. Depletion of SP1 coincided with repression of multiple glycolysis and PPP genes. Most importantly, SP1 knockdown increased ROS in Nutlin-treated cells and sensitized the cells to apoptosis. Based on these results, we propose that high levels of MDM2 in MDM2-amplifed cells treated with Nutlin cause degradation of SP1, and this results in repression of PPP genes and PPP activity, increased ROS, and death. This offers a potential explanation for why MDM2-amplified cells are especially sensitive to Nutlin-induced apoptosis. A final question is the source of ROS in Nutlin-treated and MDM2-amplifed cells. We speculate that ROS is likely derived from the mitochondria, but this remains to be determined.
MDM2 antagonists (e.g. Nutlin) have been developed as potential therapeutics against p53 wild-type cancers. However, some cancer cells undergo apoptosis as their primary response to Nutlin whereas others undergo cell cycle arrest as their primary response. While cells that undergo apoptosis will be permanently eliminated from the proliferating pool, arrested cells have the potential for regrowth after Nutlin removal. Our data suggest that targeting the PPP could be an effective way to induce apoptosis in Nutlin-treated cells that would otherwise undergo arrest, in this way expanding the breadth of cells susceptible to Nutlin-induced apoptosis.
Materials and methods
Cells and reagents
SJSA1, U2OS, and A549 cells were obtained from ATCC. MHM cells were kindly provided by Dr Ola Myklebost, Norwegian Radium Hospital. SJSA1, MHM, and A549 cells were grown in RPMI medium, U2OS in DMEM medium with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were plated 48 h before being treated with Nutlin (Sigma-Aldrich) at the indicated concentration. AZD8055, MK2206, OSI906, and rapamycin were obtained from Selleck Chemicals. Oligomycin, glucose, and 2-DG were prepared following manufacturer’s instructions that were supplied in the XF glycolysis test kit (Seahorse Bioscience).
Immunoblotting
Whole-cell extracts were prepared by scraping cells in lysis buffer (150 mM NaCl, 5 mM EDTA, 0.5% NP40, 50 mM Tris, pH 7.5), resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride membranes (Thermo Fisher Scientific). Antibodies to pAKT (T308, S473), pan AKT, pS6K (T389), S6K, pAMPK-α (T172), AMPK-α, pNDRG1 (T346), NDRG1, pRictor (T1135), Rictor, G6PD, SP1, and TSC2 were from Cell Signaling; β-actin and p53 (DO-1) were from Santa Cruz. Primary antibodies were detected with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Life Technologies), using Clarity chemiluminescence (BIO-RAD).
Flow cytometry
For cell cycle analysis, cells were harvested and fixed in 25% ethanol overnight. The cells were then stained with propidium iodide (25 μg/ml, Calbiochem). Flow cytometry analysis was performed on a Gallios™ Flow Cytometer (Beckman Coulter), analyzed with FlowJo 10 (Treestar Inc.). For each sample, 10000 events were collected.
siRNA-mediated transient knockdown
TSC2 siRNA, SP1 siRNA, G6PD siRNA, TIGAR siRNA (On-target plus smart pool), and control siRNA (On-target plus siControl non-targeting pool) were purchased from GE Dharmacon and were transfected according to the manufacturer’s guidelines using DharmaFECT I reagent.
shRNA-mediated stable knockdown
The lentiviral pLVUT-KRAB p53 shRNA was described (a generous gift from Dr Patrick Aebischer); the lentiviral packaging and envelop vectors psPAX2 and pMD2G (Addgene plasmid 12260 and 12259 deposited by Dr Didier Trono) and the pLKO-control shRNA (Addgene plasmid 1864 deposited by Dr David M. Sabatini) were obtained from Addgene plasmid repository. Lentiviral supernatants for the expression of shRNAs were generated from 293FT cells using psPAX2 and pMD2G packaging and envelope vectors according to the OpenBiosystems protocol. MHM and A549 cells were infected to establish polyclonal lines.
RNA isolation and real-time quantitative PCR analysis
Total RNA was prepared using Total RNA Mini Kit (IBI Scientific); the first cDNA strand was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Manufacturers’ protocols were followed in each case. The PCR primers for G6PD, PFK1, PFKP, PGD, PDGL, SP1, and β-actin are listed in Supplementary Table S1. SYBR green PCR kit (Applied Biosystems) was used according to the manufacturer’s instructions. AB7300 system was used as follows: activation at 95°C; 2 min, 40 cycles of denaturation at 95°C; 15 sec and annealing/extension at 60°C; 60 sec, followed by melt analysis ramping from 60°C to 95°C. Relative gene expression was determined by the ΔΔCt method using β-actin to normalize.
Assay for glycolysis
Cells were seeded using culture media at 20000 cells/well of XF96 cell plate (Seahorse Bioscience) 48 h before the assay. On the day of the assay, the media was changed to DMEM (without serum, glucose or bicarbonate, but with 2 mM Glutamine), and incubated for 2 h before the assay in a non-CO2 incubator at 37°C. Injections of glucose (10 mM final), oligomycin (5 μM final), and 2-DG (0.1 M final) were diluted in the DMEM media and loaded onto ports A, B, and C, respectively. The machine was calibrated and the assay was performed using glycolytic stress test assay protocol as suggested by the manufacturer (Seahorse Bioscience). The assay was run in one plate with 6−12 replicates. The assay was repeated at least three times. The rate of glycolysis is reported as extracellular acidification rate, or ECAR (mpH/min), after the addition of glucose.
Quantitative measurement of intracellular ROS
Cells were loaded with DHE (2 μM in PBS) for 30 min and then trypsinized. Collected cells were washed with PBS for three times, an equal number of cells were transferred to a 96-well plate, and analyzed with a BioTekMx microplate reader (excitation/emission of 510/595 nm).
Quantification of intracellular NADPH
Cells were lysed and cellular NADPH was quantified according to the manufacturer’s guidelines using NADP/NADPH Quantification Kit (Sigma-Altrich).
Glucose flux analysis
Flux studies were described previously (Ganapathy et al., 2014). In brief, cells were treated with Nutlin for a 12-h period and washed thoroughly with glucose-free medium and incubated the cells with medium containing 10 mM 1:1 mixture of D-[1,2-13C] glucose and unlabeled D-glucose for 15 min. Metabolites were extracted on dry ice with 80% methanol. The metabolites were dried under nitrogen and resuspended in 20 μl of water for LC–MS analysis.
Statistical analysis
One-way analysis of variance (ANOVA) and Student’s t-test were used to determine the statistical significance of differences among experimental groups. Student’s t-test was used to determine the statistical significance between control and experimental groups.
Supplementary Material
Funding
This work was supported by the National Cancer Institute grant 1 R21 CA185036-01A1 (to C.G.M.) and by a grant from the Swim Across America (to C.G.M.) The studies were also supported by grants from NIH, National Heart, Lung and Blood Institute (R01 HL132871 and R01 HL057832 to L.A.B.).
Conflict of interest: none declared.
Author contributions: L.D. contributed to design of experiments, acquisition of data, interpretation of data. R.E.P. contributed to acquisition of data. L.C. contributed to acquisition of data. L.A.B. contributed to design of experiments. C.G.M. contributed to design of experiments, interpretation of data, and drafting the article.
References
- Archer M.C. (2012). Role of sp transcription factors in the regulation of cancer cell metabolism. Genes Cancer 2, 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K., Tsuruta A., Selak M.A., et al. (2006). TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120. [DOI] [PubMed] [Google Scholar]
- Budanov A.V., and Karin M. (2008). p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davaadelger B., Duan L., Perez R.E., et al. (2016). Crosstalk between the IGF-1R/AKT/mTORC1 pathway and the tumor suppressors p53 and p27 determines cisplatin sensitivity and limits the effectiveness of an IGF-1R pathway inhibitor. Oncotarget 7, 27511–27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Perez R.E., Davaadelger B., et al. (2015). p53-regulated autophagy is controlled by glycolysis and determines cell fate. Oncotarget 6, 23135–23156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., and Levine A.J. (2010). The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 20, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze A., Ferrante M.I., Fusco F., et al. (1998). Molecular anatomy of the human glucose 6-phosphate dehydrogenase core promoter. FEBS Lett. 437, 313–318. [DOI] [PubMed] [Google Scholar]
- Ganapathy S., Xiao S., Yang M., et al. (2014). A low-dose arsenic-induced p53 protein-mediated metabolic mechanism of radiotherapy protection. J. Biol. Chem. 289, 5340–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez J.M., and Alessi D.R. (2008). mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., et al. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Huang J., Dibble C.C., Matsuzaki M., et al. (2008). The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell. Biol. 28, 4104–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., and Manning B.D. (2008). The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 412, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., and Manning B.D. (2009). A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 37, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., and McLachlan A. (1994). Novel clustering of Sp1 transcription factor binding sites at the transcription initiation site of the human muscle phosphofructokinase P1 promoter. Nucleic Acids Res. 22, 5085–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C.F., and Vousden K.H. (2015). p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405. [DOI] [PubMed] [Google Scholar]
- Kruse J.P., and Gu W. (2009). Modes of p53 regulation. Cell 137, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N., and Vousden K.H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Laptenko O., and Prives C. (2006). Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 13, 951–961. [DOI] [PubMed] [Google Scholar]
- Li H., Zhang Y., Strose A., et al. (2014). Integrated high-throughput analysis identifies Sp1 as a crucial determinant of p53-mediated apoptosis. Cell Death Differ. 21, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Kon N., Jiang L., et al. (2012). Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M., Thomas S.D., Islam A., et al. (2012). c-Myc and cancer metabolism. Clin. Cancer Res. 18, 5546–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J., Jung D., Wilczynski S., et al. (1998). The MDM2 gene amplification database. Nucleic Acids Res. 26, 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly K.E., Rojo F., She Q.B., et al. (2006). mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleaga C., Welten S., Belloc A., et al. (2012). Identification of novel Sp1 targets involved in proliferation and cancer by functional genomics. Biochem. Pharmacol. 84, 1581–1591. [DOI] [PubMed] [Google Scholar]
- Oliner J.D., Saiki A.Y., and Caenepeel S. (2016). The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb. Perspect. Med. 6, a026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra K.C., and Hay N. (2014). The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. (2006). Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer 6, 663–673. [DOI] [PubMed] [Google Scholar]
- Singh S., Ramamoorthy M., Vaughan C., et al. (2013). Human oncoprotein MDM2 activates the Akt signaling pathway through an interaction with the repressor element-1 silencing transcription factor conferring a survival advantage to cancer cells. Cell Death Differ. 20, 558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., Ouyang G., and Bao S. (2005). The activation of Akt/PKB signaling pathway and cell survival. J. Cell. Mol. Med. 9, 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C., Graves B., Packman K., et al. (2013). MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 73, 2587–2597. [DOI] [PubMed] [Google Scholar]
- Tovar C., Rosinski J., Filipovic Z., et al. (2006). Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl Acad. Sci. USA 103, 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Vousden K.H., and Prives C. (2009). Blinded by the light: the growing complexity of p53. Cell 137, 413–431. [DOI] [PubMed] [Google Scholar]
- Wang S.J., and Gu W. (2014). To be, or not to be: functional dilemma of p53 metabolic regulation. Curr. Opin. Oncol. 26, 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P.S., and Thompson C.B. (2012. a). Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P.S., and Thompson C.B. (2012. b). Signaling in control of cell growth and metabolism. Cold Spring Harb. Perspect. Biol. 4, a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawacka-Pankau J., Grinkevich V.V., Hunten S., et al. (2011). Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J. Biol. Chem. 286, 41600–41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.