Figure 2.
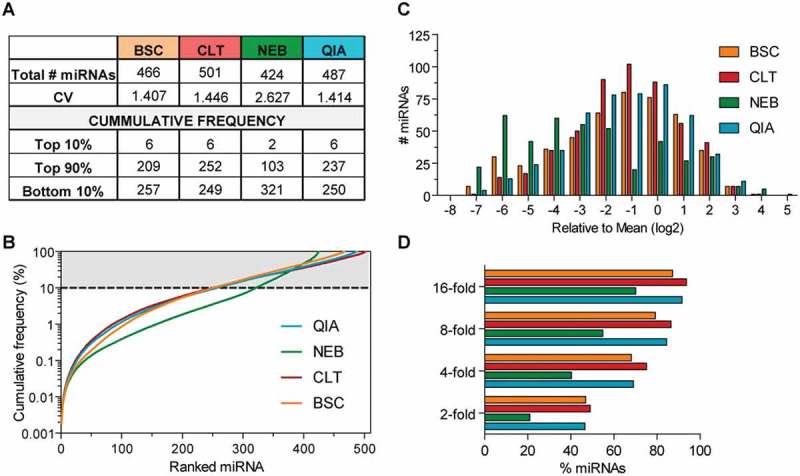
Diversity and dynamic range of sRNA-seq libraries utilising a synthetic miRNA reference.
Parallel testing of the sRNA-seq library preparation methodologies against the miRXplore universal reference (Miltenyi Biotec) library containing 539 synthetic miRNAs of human origin (n = 2 technical replicates). (a) Table summarising sensitivity and library complexity. The number of detected microRNAs with > 10 reads (using mean counts of the replicates) and the coefficient of variation (CV) were determined. (b) Cumulative frequency plot of the mean miRNA counts of the replicates for each methodology. The number of miRNAs that comprise the top 10%, the top 90% (where top means most abundant), and the bottom 10% of all miRNA reads (least abundant) for each methodology are listed in the table (a). The grey shaded area corresponds to miRNAs accounting for the top 90% of reads. (c) Histogram showing the log2 transformed ratio of individual miRNA count versus the mean count of all miRNAs. (d) Bar chart depicting the percentage of miRNAs within x-fold of the mean.
