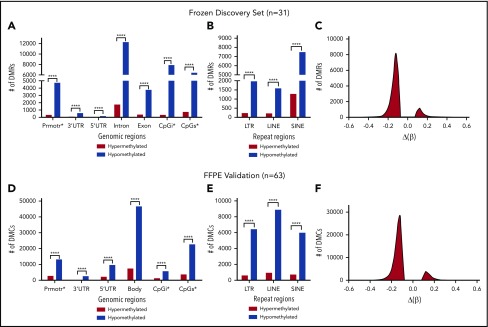
Figure 4.
ALCLs with DUSP22 rearrangements are hypomethylated. (A) RRBS of DNA extracted from frozen tissue in the 31-sample ALCL discovery set. DMRs reflect comparison of ALCLs with vs without DUSP22 rearrangements and show marked hypomethylation in DUSP22-rearranged ALCLs across all types of genomic regions, including promoters, 3′ and 5′ untranslated regions (UTRs), introns, exons, CpG islands (CpGi), and CpG shores (CpGs). (B) Hypomethylation in DUSP22-rearranged ALCLs in the discovery set involves all types of noncoding regions, including long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs). (C) Histogram showing the distribution of methylation changes [Δ(β)] for DMRs across the genome in the discovery set. (D) MethylationEPIC BeadChip array analysis of DNA extracted from FFPE tissue in an independent validation set of 63 ALCLs. Designations are similar to A, except differentially methylated CpG probes (DMCs) are shown and intron and exon data are represented together as gene bodies. DUSP22-rearranged ALCLs are hypomethylated across all types of genomic regions. (E) Hypomethylation in DUSP22-rearranged ALCLs in the validation set involves all types of noncoding regions. (F) Histogram showing the distribution of methylation changes [Δ(β)] for DMRs across the genome in the validation set. ****P ≤ .0001.