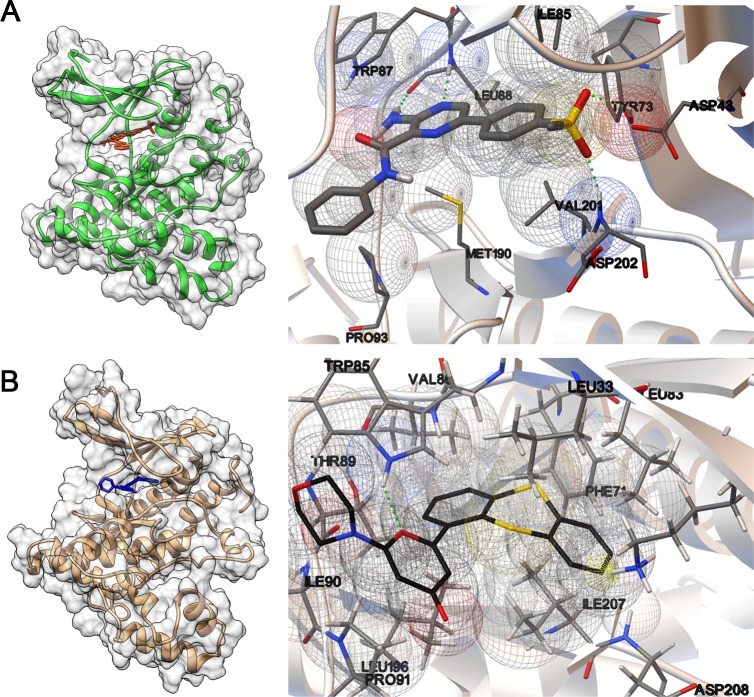
Fig 4. View of interaction site of putative LmjATR and LmjATM with the human inhibitors.
Left panels: Molecular modeling of the C-terminal region of LmjATR (A) and LmjATM (B). The sequences used for the modeling correspond to the predicted PIKK domains and range from amino acids 2863 to 3135 and 4537 to 4817 in LmjATR and LmjATM, respectively. The catalytic domains of LmjATR and LmjATM are displayed with the corresponding human inhibitors, VE-821 (ATRi—red) and KU-55933 (ATMi—blue). Right panels: View of respective docking sites. The compound structures and residue side chains predicted to participate in the interaction are displayed as sticks. ATRi is shown as thicker sticks and ATMi is shown as black sticks. Green dotted lines indicate hydrogen bonds, wireframe yellow cone indicates pi-cation interaction and wireframe spheres indicate Van der Waals interactions. Protein tertiary structure prediction was performed with ROBETTA and docking was performed with Autodock v. 4.2.6. The interactions of proteins and compounds were visualized with Chimera v. 1.11.2 (Left side) [http://www.rbvi.ucsf.edu/chimera/] and Autodock Tools v. 1.5.6 (right side) [http://autodock.scripps.edu/resources/adt].