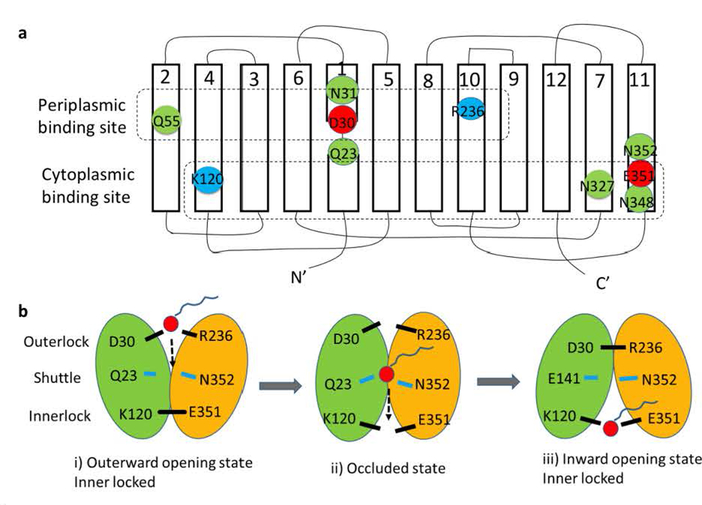
Fig. 4. Hypothetical model of the LplT-mediated lysophospholipid transport mechanism.
Panel a, Secondary structure of LplT showing the locations of the conserved residues in the putative substrate translocation pathway. The transmembrane topology diagram was rendered based on a structural homolog model of LplT from Klebsiella pneumonia generated previously (16), showing twelve transmembrane helices (1–12). Individual amino acids are represented by circles, with negatively charged residues in red, positively charged residues in blue, and other polar residues in green. The residues predicted to form the periplasmic binding site, the shuttle site and the cytoplasmic binding site are surrounded by dashed lines, respectively. Panel b, a hypothetical rocker-switch model for LplT-mediated LPL transport. The N-terminal domain formed by helices 1–6 and C-terminal domain formed by helices 7–12 of LplT are colored in green and yellow, respectively. The following three-step mechanism is proposed for LplT transport: i) an outward-facing conformation is stabilized by the outerlock formed by residues K120 and E351, which allows LPL access to the periplasmic binding site; ii) substrate binding induces a protein conformational change to open the innerlock to form an occluded state, allowing the substrate to pass via the shuttle site in the middle of the membrane towards the cytoplasmic binding site; iii) an inward-opening state is induced by outerlock formation of residues D30 and R236, which allows the substrate to be released from the cytoplasmic site to the inner leaflet of the membrane.