Figure 5.
Structural and Functional Analysis of the CENP-OPQUR Complex
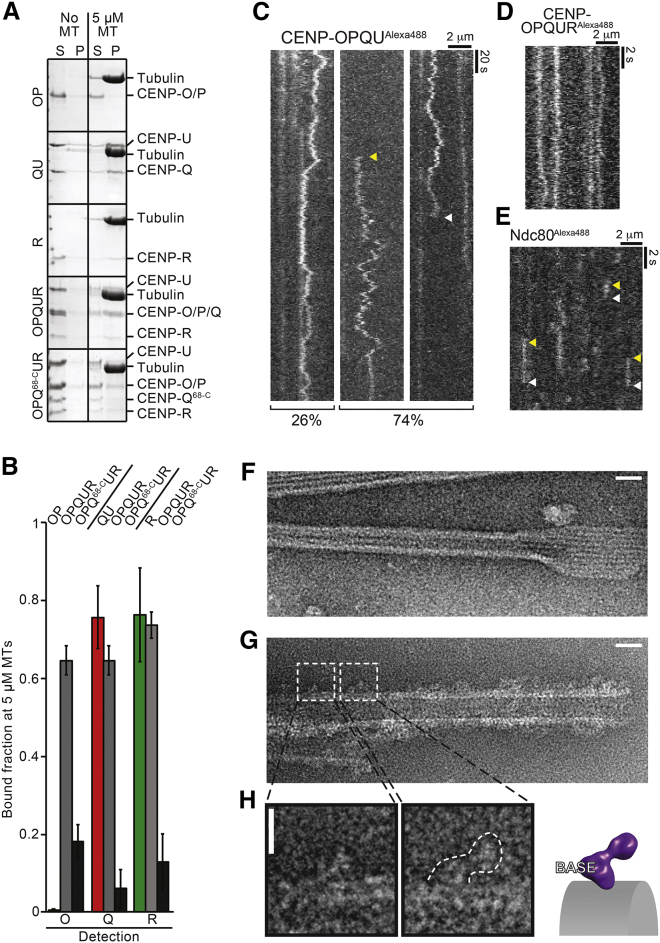
(A) Representative SDS-PAGE of microtubule co-sedimentation assays with Taxol-stabilized microtubules and the indicated proteins. CENP-OP did not bind microtubules unless it was combined with CENP-QU and CENP-R. CENP-R bound microtubules autonomously. CENP-OPQ68-CUR does not sediment with microtubules.
(B) Quantification of experiments in (A). Error bars are standard deviations calculated from three technical replicas.
(C) Kymographs of CENP-OPQU particles labelled with Alexa488 imaged on Taxol-stabilized microtubules by TIRF microscopy (n = 279). Left side shows example of particles (26% of events) that remain bound for the full duration of the video; right sides (74% of events) show examples of particles landing (yellow arrow; n = 18) or unbinding (white arrow; n = 169).
(D) Kymographs of CENP-OPQUR particles labeled with Alexa488 at higher temporal resolution (100 ms/fr).
(E) Kymographs of Alexa488-labeled NDC80 particles labeled with Alexa488 showing binding and unbinding of the Alexa488-labeled NDC800 (100 mn/fr).
(F) Representative electron micrographs of negative-stained Taxol-stabilized microtubules. Scale, 100 nm.
(G) As in (F), with added CENP-OPQUR. Scale, 100 nm.
(H) The outline of the complex recognizable on the microtubule surface suggests that the microtubule-binding moiety is in the base domain. Scale, 10 nm. For additional examples, see Figure S6F.