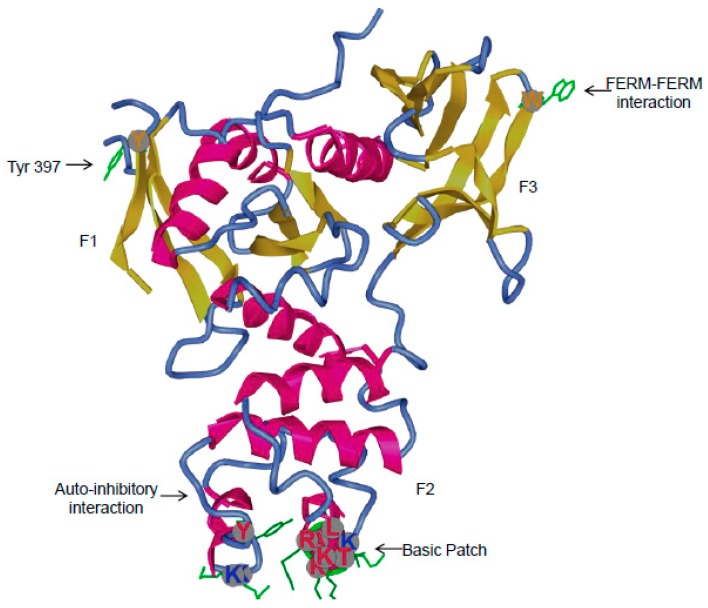
Figure 2.
Structure of the FERM domain of FAK from the protein databank (accession code 2AL6). The FERM domain displays three lobes, F1, F2 and F3. The structure includes the Tyr397 auto-phosphorylation site which is located between the FERM and kinase domain. In the F2 lobe, residues belonging to basic patches important for FAK activation are highlighted and their side chains coloured in green. The auto-inhibitory interaction implicates residues from the F2 lobe of the FERM domain and C lobe from the kinase domain. The Trp266 implicated in FERM-FERM interaction necessary for FAK dimerization is also highlighted. Proteins associating with the FERM domain are shown in the table.