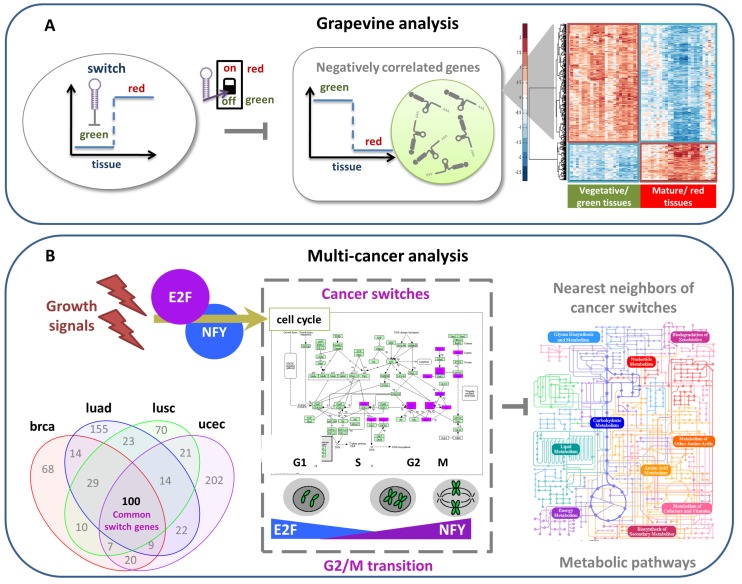
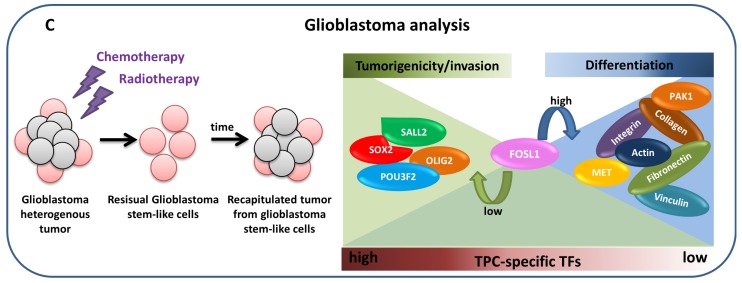
Figure 6.
SWIM applications in (A) grapevine analysis [29], (B) multi-cancers analysis [28], and (C) glioblastoma analysis [30]. (A) A sketch of the switch gene regulation mechanism in grapevine. During the vegetative/green phase of organ development, switch genes are repressed by miRNAs and vegetative genes are expressed. In the transition to the mature/red phase, these miRNAs are deactivated, the switch genes are expressed and their anti-correlated vegetative genes are turned off. The heat map shows the transcription level of positively and negatively correlated genes with a typical switch, where expression values increase from blue to red. (B) A sketch of the switch gene regulation mechanism in human cancers. SWIM extracted a set of 100 cancer-recurrent switch genes across four tumors—breast invasive carcinoma (brca), lung squamous cell carcinoma (lusc), lung adenocarcinoma (luad), uterine corpus endometrial carcinoma (ucec)—that showed a marked functional enrichment in cell cycle and specifically on the regulation of the G2-to-M transition. The promoter motif analysis suggested that two major transcription factors (namely E2F and NFY) lead to the activation of the switch gene layer of gene regulation. Activation of switch genes in these cancers seems to predominantly repress several metabolic pathways, possibly leading to the well-known metabolic rewiring characterizing cancer cells. (C) A sketch of the switch gene regulation mechanism in human glioblastoma. Glioblastoma subpopulation of self-renewing, stem-like cells has been shown to be responsible for tumor initiation, progression, resistance to treatment, and relapse. Among switch genes identified by SWIM involved in the transition from a stem-like to a differentiated phenotype of glioblastoma cells, FOSL1 stands out as a promising candidate to trigger the differentiation. On one hand, it has been found positively correlated with genes encoding proteins linked to the focal adhesion complex and extracellular matrix (ECM) receptor interaction (e.g., integrins, collagen, and signaling proteins). Conversely, it is negatively regulated with well-known neurodevelopmental transcription factors (TFs) specific of stem-like identity, including the core set of OLIG2, POU3F2, SALL2, SOX2 [70]. Thus, it could be considered as putative controller of stem-like cell differentiation process by repressing the core set of neurodevelopmental TFs and by modulating the equilibrium between cell adhesion and migration.