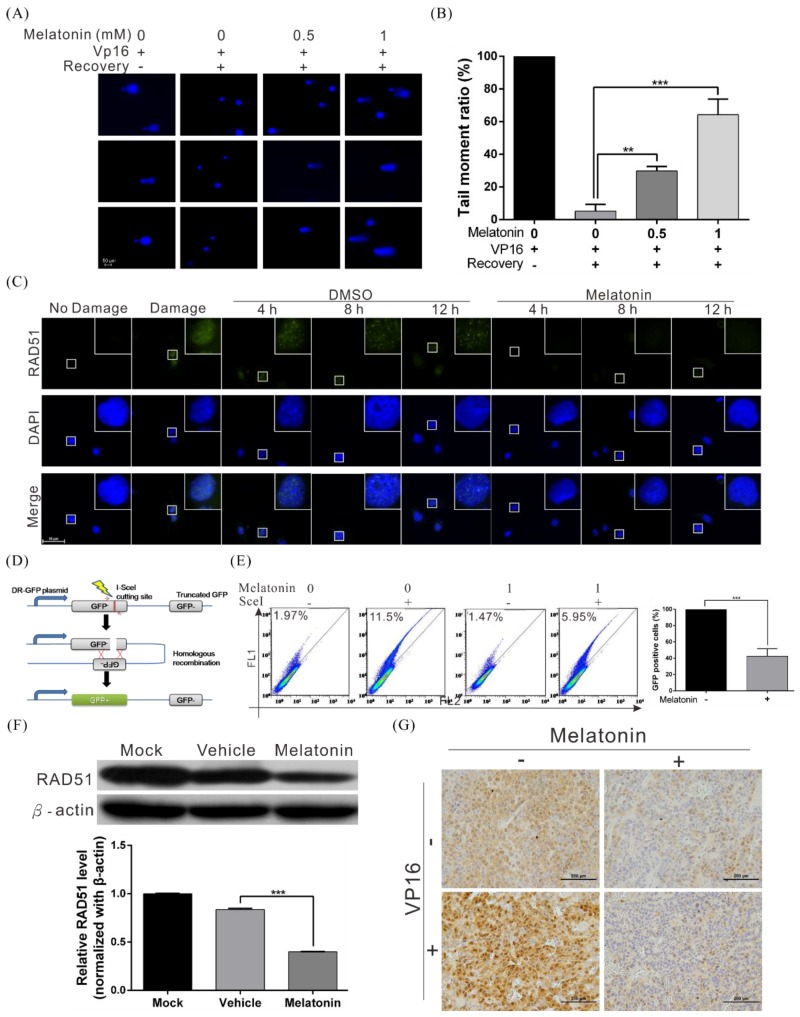
Figure 4.
Melatonin suppressed the DNA repair capacity of HCC cells by inhibiting RAD51 expression. (A) A comet assay shows DNA repair activity after treatment with melatonin. Huh7 cells were treated with 200 µM etoposide (VP16) for 1 h, followed by treatment with different concentrations of melatonin in etoposide-free medium for 4 h. The cells were harvested and subjected to comet assays to detect DNA repair activity. The rows of panels present the results of three individual experiments. Quantification of cell repair activity is shown in (B). p < 0.01 (**), p < 0.001 (***). (C) Huh7 cells were treated with 200 μM etoposide (VP16) for 1 h to induce DNA damage, followed by recovery in medium with or without 1 mM melatonin. Cells were processed for immunofluorescence staining at various time points to detect the formation of Rad51 foci (green). Nuclei were stained with DAPI (blue). (D) A schematic representing the principle of the HR reporter assay. The DR-GFP system, which contains two mutated GFP genes (termed GFP−) was applied to detect HR repair in human cancer cells. As the left GFP- gene contains an I-SceI endonuclease site, expression of I-SceI leads to a DSB that can be repaired by HR using the homologous region in the truncated GFP gene. The complete HR results in expression of a functional GFP (GFP+) that can be detected by flow cytometry. (E) The results of flow cytometry indicate that melatonin significantly inhibited HR in HCC cells. (F) Western blot analysis of RAD51 in Huh7 cells treated with or without 1 mM melatonin for 48 h; β-actin was included as an internal control (upper panel). Quantitative results are shown in the lower panel. (G) Downregulation of RAD51 in mice xenografts treated with melatonin, as examined by immunohistochemistry.