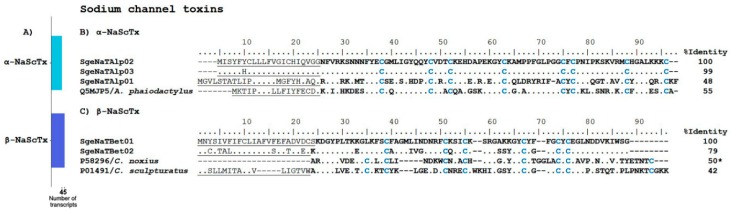
Figure 3.
Putative Na+ channel-specific toxins encoded by S. gertschi transcripts. (A) Number of different transcripts putatively coding for α-NaScTx and β-NaScTx. The number of transcripts is the number of unique sequences that putatively encodes different proteins of this subcategory. (B) The translated transcripts putatively coding for ion channel toxins similar to phaiodotoxin. (C) Transcript-derived partial peptide sequences coding for β-NaScTx aligned to their closest matches by BLAST. Dots indicate sequence identity; dashes indicate gaps. The predicted signal peptides are underlined, the mature peptides are in bold typeface, and the cysteines are highlighted in blue. Reference proteins include their UniProt identifiers and the species’ name. The identity percentages (%I) in the alignments were calculated based on the precursor sequences, except where indicated by an asterisk (*), for which only the mature sequence was considered.