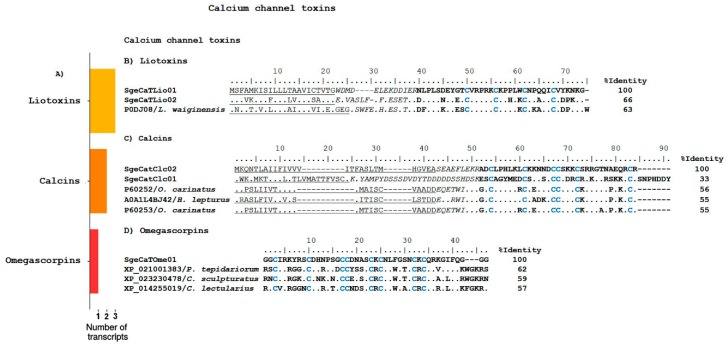
Figure 5.
Putative Ca2+ channel toxins identified by the transcriptome analysis of S. gertschi. (A) Distribution of the different transcripts putatively coding for calcins, liotoxins, and omegascorpins. The number of transcripts is the number of unique sequences that putatively encodes to the different proteins of this subcategory. (B) The CDS from the transcripts coding for putative liotoxins aligned to reference precursor sequences. (C) Transcript-derived precursor sequences coding for calcins together with previously reported similar precursor sequences. (D) Transcript-derived mature predicted sequences that putatively code for omegascorpins, aligned with their best matches according to BLAST. The predicted signal peptides are underlined, the propeptides are in italics, the mature peptides are in bold typeface, and the cysteines are highlighted in blue. Reference proteins include their UniProt identifiers and the species’ name. The identity percentages (%I) in the alignments were calculated based on the precursor sequences for the liotoxins and calcins, or the mature sequences for the omegascorpins.