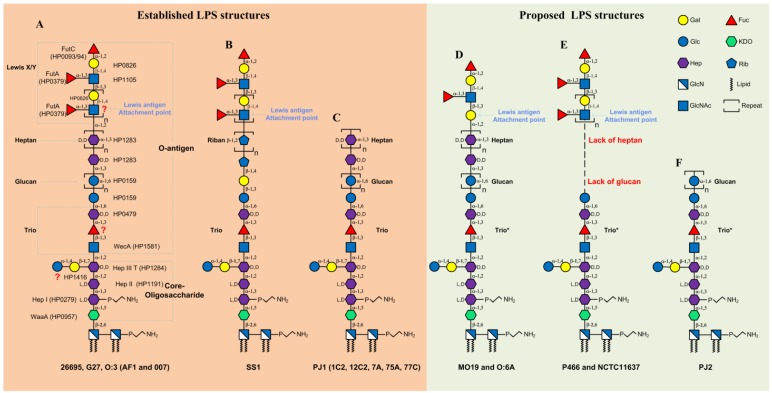
Figure 1.
Established and proposed lipopolysaccharide (LPS) structures in Western H. pylori strains. Only three established LPS structures are available (A–C). Assuming the Trio is conservatively present in H. pylori LPS, 3 additional LPS structural models can be proposed from partial structures reported in the literature (D–F). (A) LPS structures in strains 26695 [11], G27 [9], O:3 [5,10], AF1, and 007 [19]. The general architecture of the LPS structures in these strains are very similar, although the length of the glucan and heptan, the fucosylation, and number of repeats of the Lewis chains, vary between strains. The O-antigen is defined to encompass the Trio, the glucan-heptan linker and the Lewis antigens. The Lewis antigen attachment point is suggested to be a GlcNAc residue. Previously known H. pylori LPS biosynthetic enzymes are presented, whereas the unidentified enzymes are indicated by red question marks; (B) LPS structure in strain SS1 [12]. No glucan (however a single Glc residue is present after the Trio) and no heptan are present, instead, a rare riban structure (oligomer of Rib) is inserted between the core-Trio and Lewis antigen structures. The Lewis antigen is connected to the riban through a GlcNAc residue; (C) LPS structures in strains PJ1, 1C2, 12C2, 7A, 75A, and 77C [20]. LPS in these strains contains the glucan and heptan, but lacks the Lewis antigens; (D) LPS structures in strains MO19 and O:6A [5,7]. The glucan and heptan structures are present, whereas only a single Ley unit is detected in the two strains. The Lewis antigen is attached to the heptan via a Gal residue rather than the GlcNAc residue; (E) LPS structures in strains P466 [7] and NCTC11637 [14]. The glucan and heptan structures are absent in these two strains; (F) LPS structures in strain PJ2 ends with the glucan but lacks the heptan and Lewis antigens. * The Trio structure is inferred to be conservatively present in these strains.