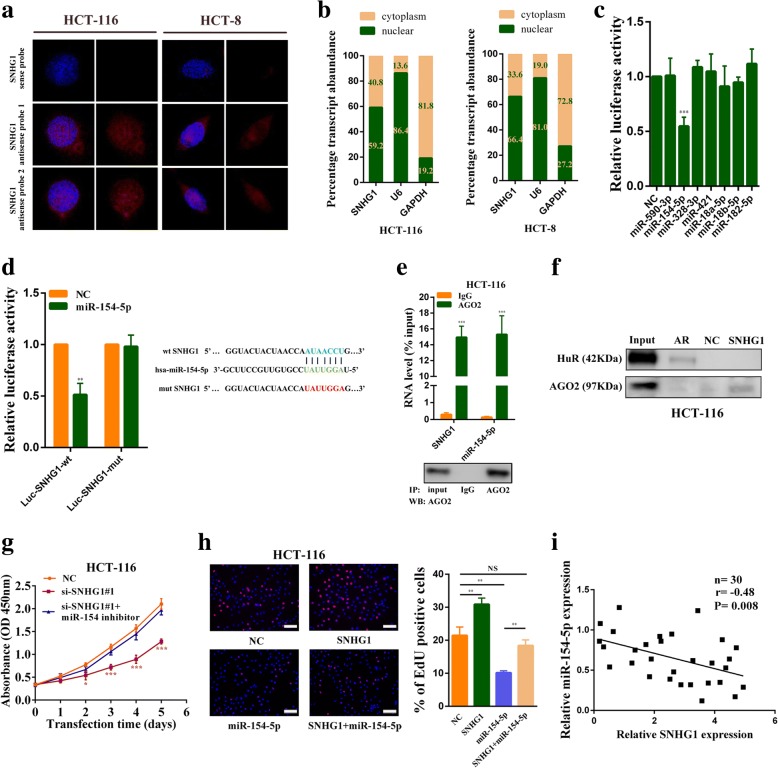
Fig. 5.
SNHG1 acts as a sponge for miR-154-5p in the cytoplasm. a Representative FISH images indicated subcellular location of SNHG1 in HCT-116 and HCT-8 cells (red). Nuclei were stained by DAPI (blue). SNHG1 sense probe was employed as a negative control. b Relative SNHG1 expression levels in nuclear and cytosolic fractions of HCT-116 and HCT-8 cells. Nuclear controls: U6; Cytosolic controls: GAPDH. c Dual luciferase reporter assays were used to determine miRNAs that directly interacted with SNHG1. Luciferase activity is presented as relative luciferase activity normalized to activity of their respective negative control. d Dual luciferase reporter assays were conducted with wild type and mutant type (putative binding sites for miR-154-5p were mutated) luciferase reporter vectors. Right panel, sequence alignment of miR-154-5p and its predicted binding sites (green) in SNHG1. Predicted miR-154-5p target sequence (blue) in SNHG1 (Luc-SNHG1-wt) and position of mutated nucleotides (red) in SNHG1 (Luc-SNHG1-mut). e RNA immunoprecipitation with an anti-Ago2 antibody was used to assess endogenous Ago2 binding to RNA in HCT-116 cells, IgG was used as the control. SNHG1 and miR-154-5p levels were determined by qRT–PCR and presented as fold enrichment in Ago2 relative to input. RIP efficiency of Ago2 protein was detected by western blot. f RNA pull-down assays were used to examine the interaction of SNHG1 and Ago2 in HCT-116 cells. g CCK-8 assays demonstrated that SNHG1 silencing inhibited HCT-116 cell growth. MiR-154-5p down-regulation rescued growth inhibition caused by SNHG1 knockdown. h EdU assays revealed that SNHG1 overexpression promotes HCT-116 cell proliferation. Co-transfecting miR-154-5p mimics with the SNHG1 plasmid abolished the increased proliferation rates. i The correlation between miR-154-5p and SNHG1 expression analyzed in 30 paired colorectal cancer samples (n = 30, r = − 0.48, P = 0.008). Scale bar = 50 μm. *P < 0.05, **P < 0.01 and ***P < 0.001