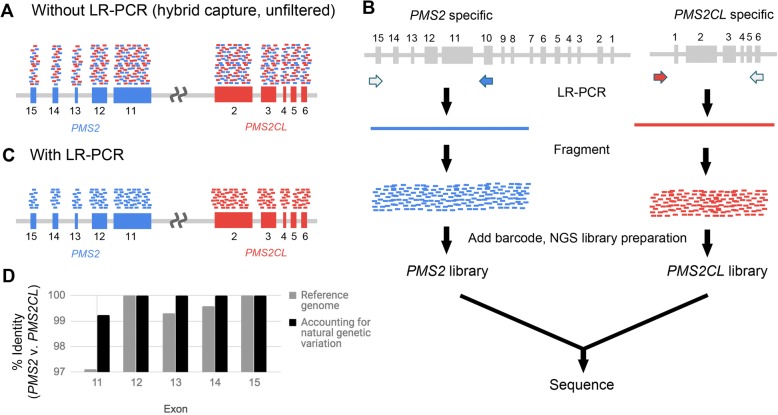
Fig. 1.
LR-PCR strategy for building a dataset of natural genetic variation in PMS2 and PMS2CL. a Short-reads from NGS hybrid-capture data that originate from the gene (blue) and pseudogene (red) align to both the gene and pseudogene due to high homology. b, c Using LR-PCR that is specific to the gene or pseudogene followed by fragmentation and barcoding (b), the resulting short NGS reads can be assigned to the gene or pseudogene (c). d Percent identity between the gene and pseudogene for PMS2 exons 11–15 based on the hg19 reference genome (gray) and after accounting for natural genetic variation obtained from LR-PCR samples (black)