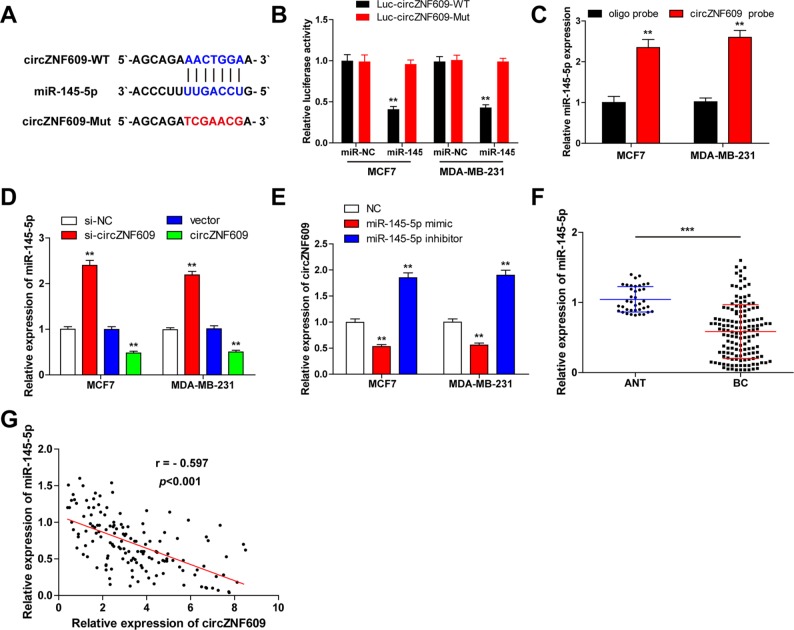
Figure 3.
CircZNF609 serves as a sponge of miR-145-5p in breast cell lines.
Notes: (A) The putative targeting site of circZNF609 and miR-145-5p was predicted by CircInteractome. (B) Luciferase activity analysis in MCF7 and MDA-MB-231 cells cotransfected with miR-145-5p mimics and pmirGLO-circZNF609-WT or pmirGLO-circZNF609-Mut vector. (C) RNA pull-down assay in MCF7 and MDA-MB-231 cells, the expression levels of miR-145-5p pulled down by circZNF609 or oligo probe were detected by qRT-PCR. (D) qRT-PCR analysis of miR-145-5p in MCF7 and MDA-MB-231 cells with circZNF609 knockdown or overexpression. (E) qRT-PCR analysis of circZNF609 in MCF7 and MDA-MB-231 cells with miR-145-5p knockdown or overexpression. (F) qRT-PCR analysis of miR-145-5p in BC tissues (n=143) and adjacent normal tissues (n=38). (G) Pearson’s correlation analysis of circZNF609 and miR-145-5p in BC tissues (n=143) (r=-0.597, P<0.001). Data were represented as mean±SD of at least three independent experiments. **P<0.01, ***P<0.001.
Abbreviations: ANT, adjacent normal tissue; BC, breast cancer; NC, negative control WT, wild type; qRT-PCR, quantitative reverse transcription PCR.