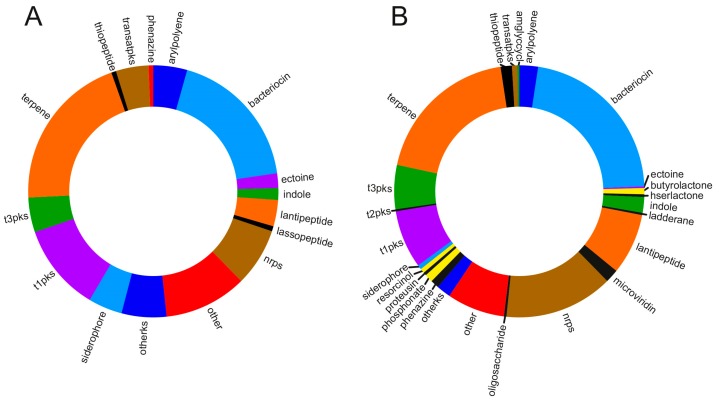
Figure 6.
Biosynthetic gene clusters in myxobacterial genomes (A) from saline environments (∑ = 181 BGCs) and (B) from terrestrial environments (∑ = 467 BGCs). For the computational analysis, we added the following genomic sequences of terrestrial myxobacteria (NCBI reference sequence accession codes in parentheses): Anaeromyxobacter dehalogenans 2CP-1 (NC_011891.1), Anaeromyxobacter dehalogenans 2CP-C (NC_007760.1), Anaeromyxobacter sp. Fw109-5 (NC_009675.1), Anaeromyxobacter sp. K (NC_011145.1), Archangium gephyra DSM 2261 (NZ_CP011509.1), Chondromyces crocatus Cm c5 (NZ_CP012159.1), Corallococcus coralloides DSM 2259 (NC_017030.1), Cystobacter fuscus DSM 52655 (NZ_CP022098.1), Melittangium boletus DSM 14713 (NZ_CP022163.1), Myxococcus fulvus 124B02 (NZ_CP006003.1), Myxococcus hansupus mixupus (NZ_CP012109.1), Myxococcus macrosporus DSM 14697 (NZ_CP022203.1), Myxococcus stipitatus DSM 14675 (NC_020126.1), Myxococcus xanthus DK 1622 (NC_008095.1), Sandaracinus amylolyticus DSM 53668 (NZ_CP011125.1), Sorangium cellulosum So ce 56 (NC_010162.1), Sorangium cellulosum So0157-2 (NC_021658.1), Stigmatella aurantiaca DW4/3-1 (NC_014623.1), Vulgatibacter incomptus DSM 27710 (NZ_CP012332.1).