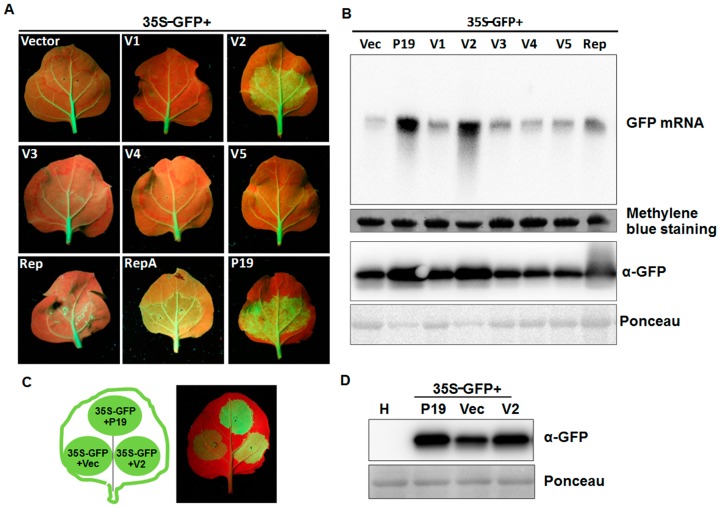
Figure 3.
Effects of MMDaV-encoded proteins on GFP local silencing. (A) N. benthamiana 16c plants were infiltrated with a mixture of Agrobacterium cultures containing 35S-GFP and pCHF3 vectors expressing individual ORFs of MMDaV, respectively. The constructs used for infiltration are indicated. N. benthamiana 16c plants infiltrated with 35S-GFP, plus the empty vector or 35S-GFP plus tomato bushy stunt virus P19 were used as negative or positive controls, respectively. Photographs were taken under UV light with a yellow filter-mounted Canon camera at 5 dpi. Similar results were obtained in three dependent experiments. At least five plants were agroinfiltrated per experiment. (B) Analysis of the GFP mRNA and protein levels in infiltrated leaf patches. 10 μg of total RNA extracted from infiltrated patches at 5 dpi were used in Northern blot analysis. The GFP mRNA was detected using a DIG-labeled GFP-specific probe. Methylene blue staining was used to visualize the loading controls for the mRNA. The expression of the GFP protein was analyzed by Western blot using a monoclonal antibody against GFP. Ponceau staining of the large subunit of Rubisco served as loading controls for the Western blot assay. (C) Suppression of local post-translational gene silencing (PTGS) in wild-type N. benthamiana plants. N. benthamiana plants were infiltrated with a mixture of Agrobacterium cultures containing constructs as indicated in the left panel. Expression of 35S-GFP with the empty vector or 35S-GFP with P19 served as negative or positive controls, respectively. Photographs were taken under UV light at 3 dpi. (D) Western blot analysis of the GFP protein levels in infiltrated N. benthamiana leaf patches using a GFP monoclonal antibody. Total soluble proteins extracted from the healthy N. benthamiana plants were used to detect the specificity of the GFP antibody. Ponceau staining of the large subunit of Rubisco protein are shown as loading controls.