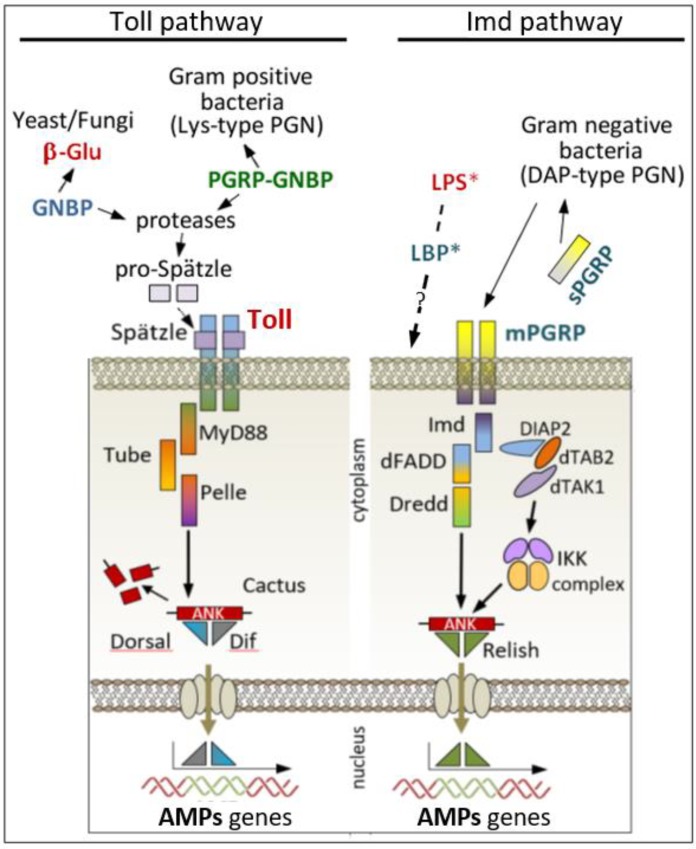
Figure 4.
Cellular PRRs: Toll and Imd pathways. (Left) a schematic view of fungine and Gram-positive bacteria stimulation of Toll pathway; the intracellular signal transduction involves adaptor proteins as MyD88, Tube and the serine/threonine protein kinase Pelle. Upon activation, Toll receptor-adaptor complex signals to a latent transcriptional factor, belonging to the NF-kB-Rel family, NF-kB factor is normally complexed to an inhibitor protein (IkB-like) named Cactus. The Toll signaling induces the degradation of Cactus and dissociation from Rel family proteins (Dorsal and Dif). Dif, a member of NF-kB-like proteins, seems to be the main transactivator factor responsible for antifungal and anti-Gram positive bacterial defenses. Dif translocates into the nucleus and switches on many AMPs (antimicrobial peptides) genes, probably several hundred, that in concert contribute to challenge microbial infection. (Right) the Imd pathway stimulated by Gram negative bacteria. The Imd gene encodes a 25 kDa protein with a death domain, the protein acts upstream of an adaptor protein named FADD; furthermore, for a full activation, a caspase-8 like protein is required (Dredd). Dredd can directly activate REL protein (Relish). Finally, Rel domain after cleavage, moves to the nucleus and activates AMPs genes. Otherwise, Relish can be phosphorylated by IKK signaling complex, which is itself activated by Tak1 (a MAP kinase) interacting downstream of Imd/FADD.