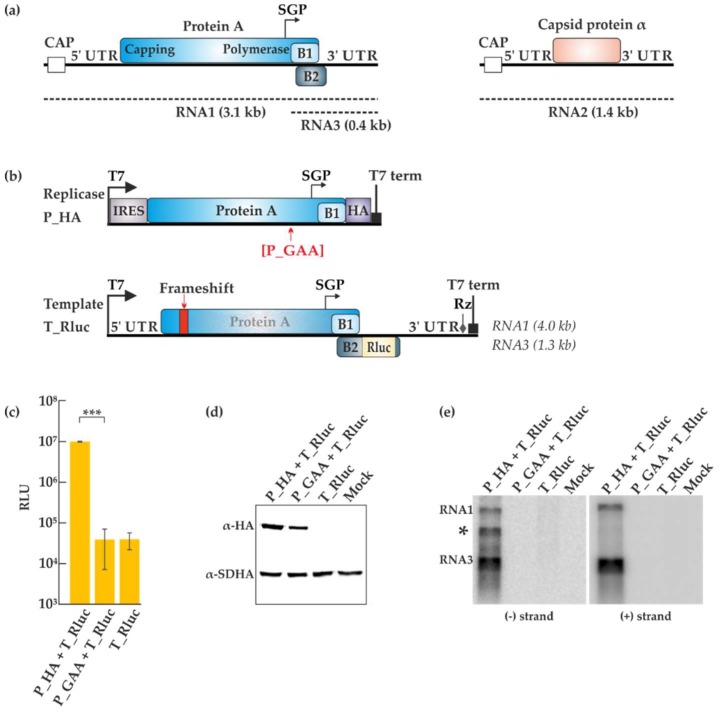
Figure 1.
Trans-replication of flock house virus (FHV). (a) Depiction of the bipartite FHV genome. Both RNA1 and RNA2 are capped, positive sense RNAs flanked by untranslated regions (UTRs). The replicase protein A is translated from RNA1. The subgenomic RNA3 arises from the subgenomic promoter (SGP) and is used for the production of the small proteins B1 and B2. RNA2 codes for the capsid protein α. The dotted lines denote the complementary negative strands generated during replication. (b) The plasmids for the trans-replication system produce RNAs from the T7 polymerase promoter, ending with the T7 terminator (T7 term) and/or ribozyme (Rz) elements. The locations for the polymerase inactivating mutation and the frameshift mutation are shown in red. (c) BSR T7/5 cells were transfected with the indicated plasmids and luciferase activity was measured 40 h post-transfection. Luciferase signals are shown as mean (corrected for mock transfected background) from two independent experiments. Error bars represent the standard deviation. *** designates p < 0.001 (Student’s t test). RLU: relative light units. (d) Replicase expression following transfections in BSR T7/5 cells was detected by Western blotting with anti-hemagglutinin (HA) antibodies. The expression of the cellular protein succinate dehydrogenase (SDHA) is shown as a control. (e) Viral RNA synthesis was analyzed by Northern blotting using probes designed to detect negative and positive strands RNAs. The asterisk marks an uncharacterized RNA product that was sometimes observed during minus strand detection. Rluc: Renilla luciferase