Fig. 1.
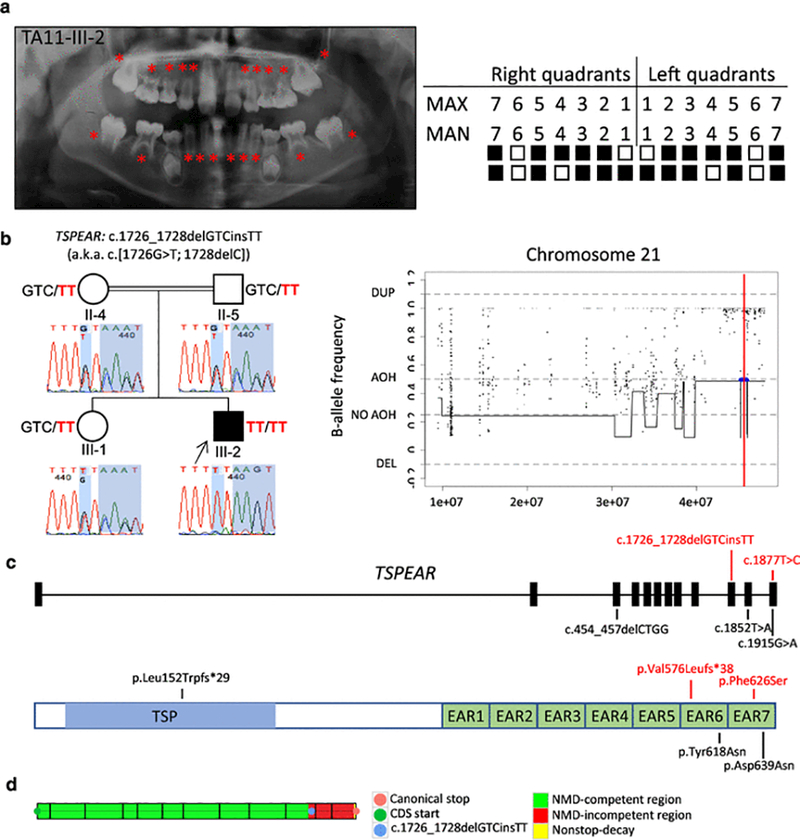
Clinical findings of TA11 family proband with tooth agenesis (TA), segregation results, absence of heterozygosity (AOH) plots, and molecular findings of patients with variants in TSPEAR. (a) TA diagnosis was confirmed through clinical and radiographic examinations for patient TA11-III-2. The panoramic radiograph was taken at 10 years 10 months. On the left, permanent teeth missing are indicated by red asterisks on the panoramic radiographs; on the right, the filled boxes represent permanent teeth missing in a schematic of the maxillary (MAX) and mandibular (MAN) arches while non-filled boxes indicate permanent teeth present excluding third molars. (b) Pedigree and genotypes of family TA11. Neighboring homozygous missense and truncating variants result in a complex allele c.1726_1728delGTCinsTT (p.Val576Leufs*38), a.k.a. c.[1726G>T; 1728delC] (red font) in TSPEAR identified in the proband (III-2, arrow), while these two variants were found in a heterozygous state in the unaffected sister (III-1) and consanguineous parents (II-4 and II-5). Sanger sequencing chromatograms demonstrate the location of identified variants (shaded in blue). AOH regions in the proband were identified by calculation of B-allele frequency from exome variant data using BafCalculator (horizontal blue line) and the position of the two variants indicated with a thin vertical red line. These two variants are located within an AOH region (~442 kb). The total calculated AOH size in the proband is 333.472 Mb. (c) Positions of the variants identified in the present study (red font) and previous studies are mapped to the 12 exons of TSPEAR. The homozygous allele c.1726_1728delGTCinsTT (in TA11) is located in exon 10, and homozygous variant c.1877T>C (in TA12) maps to exon 12; additional previously reported variants (black font) map to exons 3, 11, and 12. Below the gene structure is a schematic representation of TSPEAR protein structure. The corresponding amino acid changes are located in the thrombospondin-type laminin G (TSP) domain and two epilepsy-associated repeats (EAR) domains. (d) The complex allele c.1726_1728delGTCinsTT was predicted to escape NMD by our in-house developed tool based on the rule that PTCs within the last 50 base pairs of the penultimate exon and the last exon are hypothesized not to undergo NMD. Abbreviations: AOH, absence of heterozygosity; DEL, deletion; DUP, duplication.
