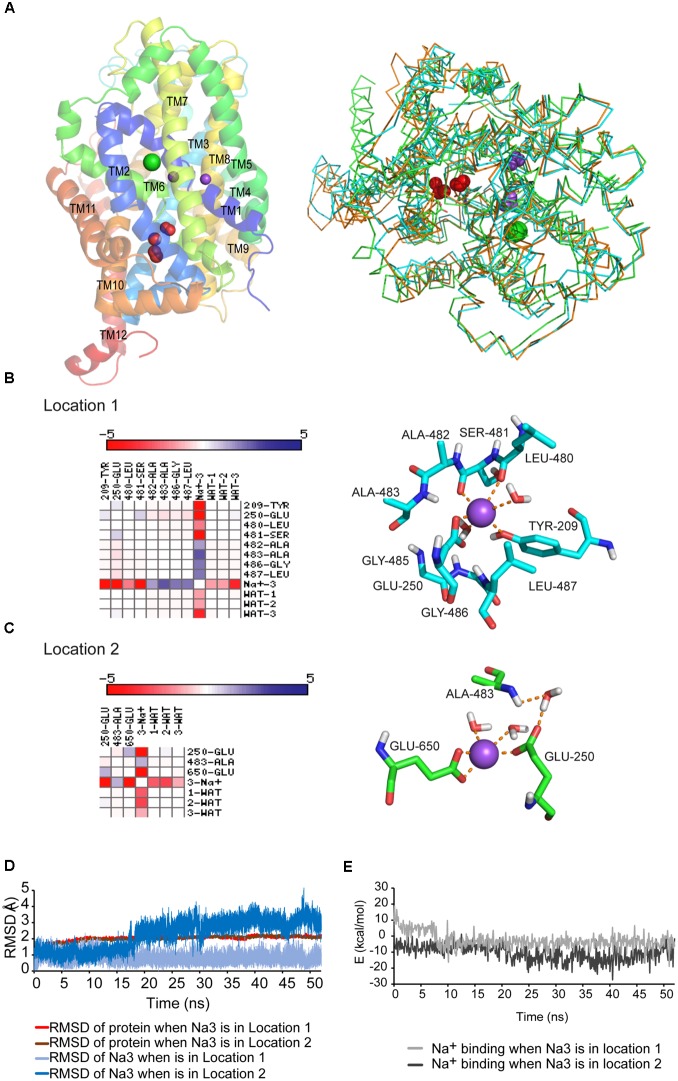
FIGURE 1.
(A) GlyT2 dDAT homology model. Left: PyMol-generated GlyT2 lateral view is shown in a cartoon representation with conserved Na1 and Na2 as purpled spheres and chloride ion as green sphere. The water molecules found in several NSS crystals (red spheres) are surrounding the unwound portion of TM6. Right: structure alignment in PyMol of GlyT2 model, LeuTAa and dDAT crystals. 90% of the LeuTAa and dDAT crystals in PDB show 2 crystallographic water molecules in locations 1 and/or 2. The three structures are shown as ribbon, GlyT2 model is colored in orange, LeuTAa in green, and dDAT in cyan. The waters are depicted as red spheres and residues Glu250 and Glu650, which bind waters, are shown in stick representation. (B) Interaction matrix and lateral view of Na3 placed in location 1. (C) Interaction matrix and lateral view of Na3 placed in location 2. Cations are shown as purple spheres. Residues are shown in stick representation. (D,E) Root-mean-square-deviations (RMSD) (D) and energy profile (E) along the 50 ns MD simulations with sodium placed in location 1 or 2.