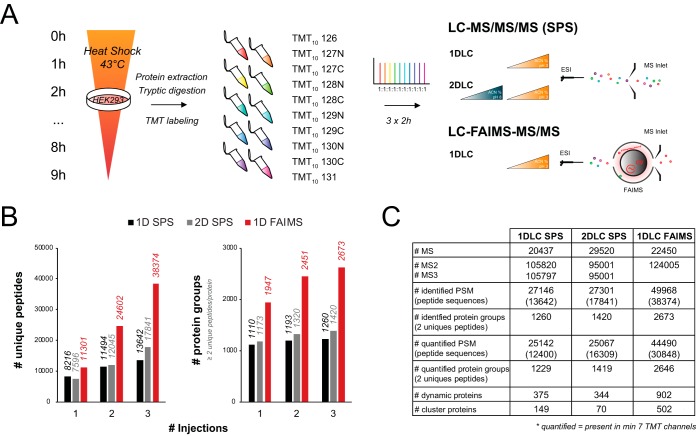
Fig. 5.
FAIMS improves TMT quantification of the human proteome. (A) TMT labeling and quantification workflow. HEK293 cells were exposed to a 43 °C heat stress for up to 9 h in 1-h increments. At each time point, cells were collected, lysed, digested with trypsin, and labeled with a different TMT10 channel. The 10 TMT-labeled samples were combined in an equimolar amount and analyzed by 2-h reversed-phase LC gradient. Triplicate injections were performed for the SPS method with and without high pH/basic reverse-phase fractionation while three different CV combinations (CV -37 V/-44 V/-51 V, CV -58 V/-65 V, and CV -72 V/-79 V/-86 V/-93 V) were used with FAIMS. (B) Cumulative number of unique peptides and protein groups identified as a function of repeat injections for 1D SPS (black), 2D SPS (gray), or CV stepping program with FAIMS (red). (C) Summary table comparing MS analysis parameters between SPS and FAIMS.