Fig. 2.
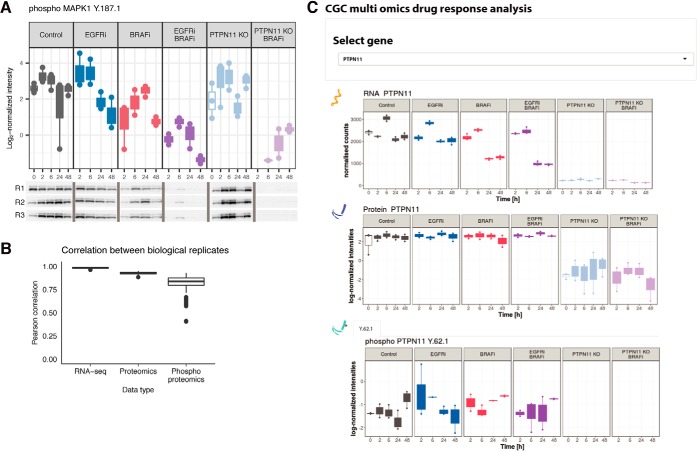
High data quality enables reliable overview of gene-level response across omics datasets. A, Label-free phosphoproteomics and phosphoWB provide similar regulation patterns. The LC-MS/MS quality control was assessed by quantification of ERK phosphorylation (top panel) in all three biological replicates (R1, R2 and R3), and subsequent validation was done via western blots by using pERK1/2 (bottom panel) which confirmed down-regulation of pERK1 on Y187 as measured by mass spectrometry. B, Quality analysis of reproducibility at each omics level. Replicate consistency was assessed by inter-replicate Pearson correlation. In the phosphoproteomic analysis, selection of technical replicates involved discarding the poorest replicates before quantification. Median of technical replicates was performed for both phospho- and proteomics dataset, and the resulting processed data were further filtered to only include data that was quantified in at least one time point of any condition. Analysis indicates good median correlation (R > 0.8) among the three biological replicates for all three omics datasets. C, Cross-omics Graphical User Interface output. The GUI enables exploration of the multi-omics data for specific genes under all the tested conditions. Users can select a measured gene of interest from a dropdown menu, and visualize complete data at the transcript, protein, and phosphosite level, over the time-course and over all experimental conditions. An example output for the gene PTPN11 is shown.