Abstract
Significance: Various autoimmune syndromes are characterized by abnormalities found at the level of tissues and cells, as well as by microenvironmental influences, such as reactive oxygen species (ROS), that alter intracellular metabolism and protein expression. Moreover, the convergence of genetic, epigenetic, and even environmental influences can result in B and T lymphocyte autoimmunity and tissue pathology.
Recent Advances: This review describes how oxidative stress to cells and tissues may alter post-translational protein modifications, both directly and indirectly, as well as potentially lead to aberrant gene expression. For example, it has been clearly observed in many systems how oxidative stress directly amplifies carbonyl protein modifications. However, ROS also lead to a number of nonenzymatic spontaneous modifications including deamidation and isoaspartate modification as well as to enzyme-mediated citrullination of self-proteins. ROS have direct effects on DNA methylation, leading to influences in gene expression, chromosome inactivation, and the silencing of genetic elements. Finally, ROS can alter many other cellular pathways, including the initiation of apoptosis and NETosis, triggering the release of modified intracellular autoantigens.
Critical Issues: This review will detail specific post-translational protein modifications, the pathways that control autoimmunity to modified self-proteins, and how products of ROS may be important biomarkers of tissue pathogenesis.
Future Directions: A clear understanding of the many pathways affected by ROS will lead to potential therapeutic manipulations to alter the onset and/or progression of autoimmune disease.
Keywords: : post-translational modification, carbonylation, citrullination, autoantigens, autoimmunity, type 1 diabetes
Introduction
At its most basic level, autoimmunity is initiated as the errant product of both cells and soluble compounds (cytokines, other soluble factors, as well as altered self-proteins), leading to specific recognition and robust binding to self-proteins and/or tissues leading to immune-mediated tissue pathology. In addition, various genome-wide association studies now conducted in virtually all autoimmune syndromes have identified specific heritable genetic traits for individuals at risk for autoimmunity. However, autoimmunity, and type 1 diabetes (T1D) in particular, cannot be entirely explained by a defined group of genes. For example, many nonheritable or stochastic factors such as poorly defined influences of our environment as well as epigenetics also control the onset and/or progression of T1D, recently reviewed in Ref. (153). The assembly of manuscripts in the present issue of Antioxidants and Redox Signaling examines the features of T1D autoimmunity linked to oxidative tissue environments. Reactive oxygen species (ROS), including hydrogen radicals (OH.), superoxide anion (O2.−), and hydrogen peroxide (H2O2), are a result of dynamic balance with natural anti-oxidant cellular products that control their concentrations and biological effects. These anti-oxidants include catalase, superoxide dismutase, peroxiredoxins, glutathione peroxidase, as well as other neutralizing small-molecule substances, vitamins E and C.
Although not elaborated within this review, the sources of ROS at sites of tissue autoimmunity are many, including the invasion of activated immune phagocytic cells (neutrophils, macrophages, and dendritic cells) that are undeniably critical to the onset, progression, and tissue pathology of T1D as well as many other autoimmune syndromes. Though ROS-mediated cellular stress induces many protein post-translational modifications (PTMs) [reviewed in Ref. (121)], this work will focus on those specific pathways that are relevant in inflammatory autoimmune syndromes.
“Oxidative stress” can trigger direct modification of certain proteins, or protein motifs, or, alternatively, as secondary modifications due to indirect metabolic pathways affected by ROS. These secondary effects include the role of ROS in apoptosis, the generation of neutrophil extracellular traps (NETs; NETosis), and intracellular metabolic pathways, all of which may affect the outcome of autoimmune responses and/or inflammatory tissue pathology. Affected proteins may be altered in solubility, in their ability to be digested or cleared, or altered in immunogenicity. As illustrated in this review, oxidation can provoke a number of cellular changes at both the DNA and protein level, the latter of which includes both spontaneous and enzyme-mediated modifications to self proteins that are relevant biomarkers of tissue pathology and autoimmunity in T1D. Herein, we will describe elements of the initiation and progression of T1D immunity that are influenced by oxidative pathways, including various post-translational protein modifications, as well as the role of oxidation at the level of DNA transcription and translation.
Post-translational modifications of self-proteins in autoimmunity
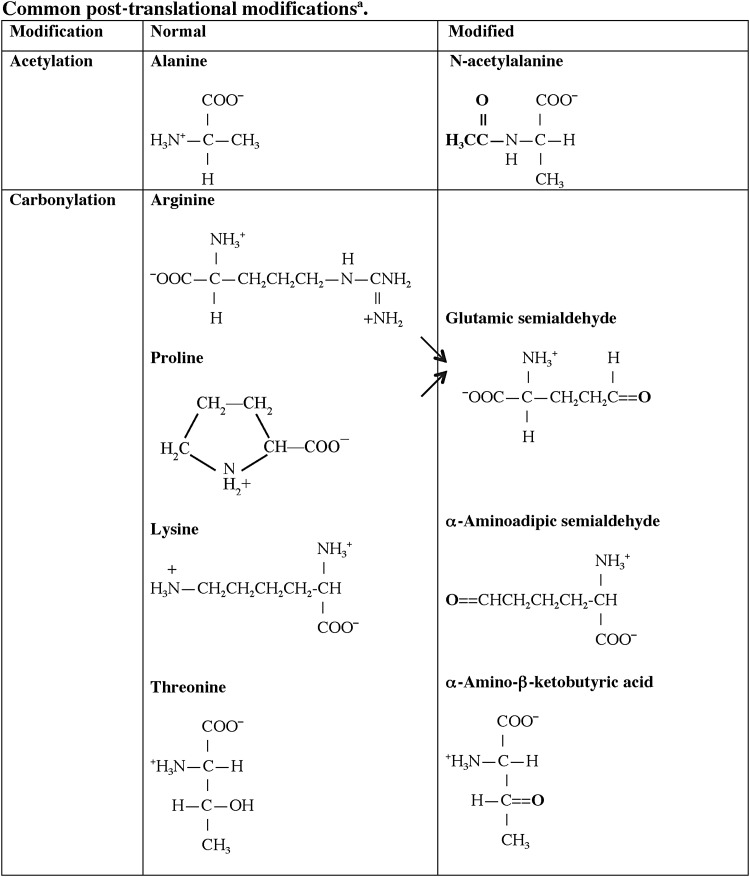
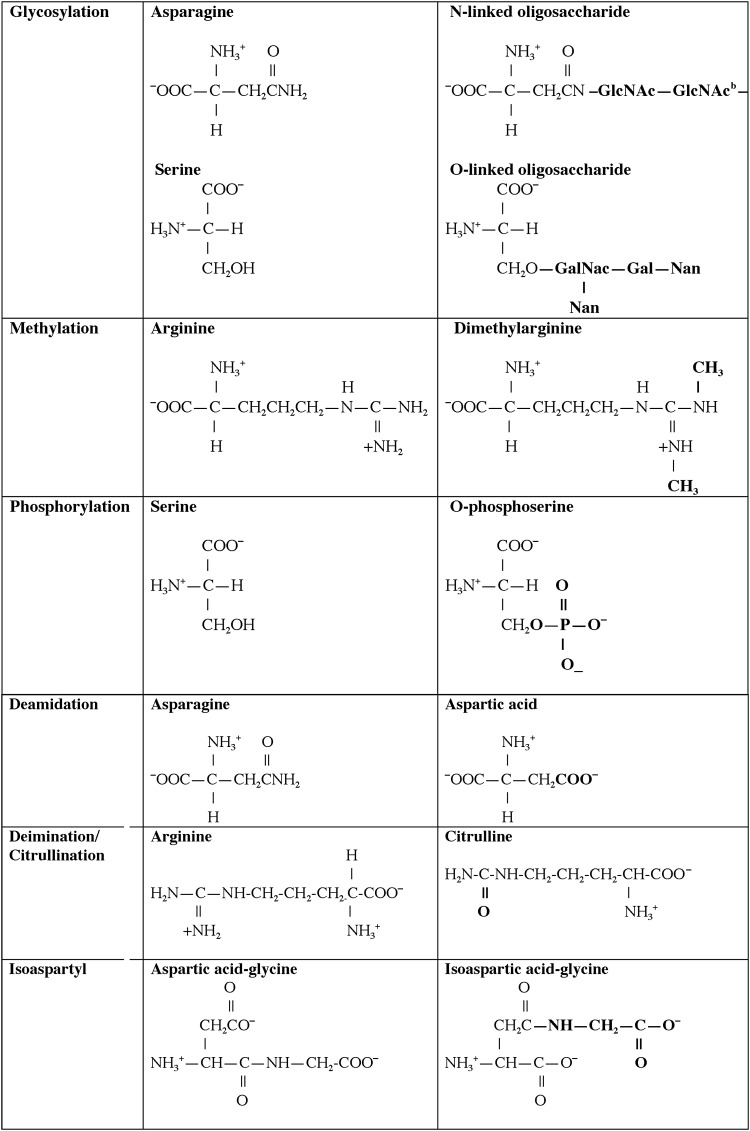
One central role of the immune system is to differentiate responses of the “self” from the “non-self” proteome. A variety of regulatory pathways are in place to deplete the host of lymphocytes that respond vigorously to self-antigens that are present in secondary lymphoid organs, specifically the bone marrow and thymus. Although many self-proteins are found in the thymus (24), the PTMs of self-antigens (Tables 1 and 2) can create a novel autoantigenic proteome to which lymphocytes have never been exposed in either the thymus or peripheral lymphoid compartments. This concept, previously described as “autoantigenesis,” is a term described to proteins that “evolve” and acquire PTMs over the course of disease and provoke B and T cell autoimmunity leading to tissue pathology (31). This phenomenon is characteristic of many autoimmune diseases such as rheumatoid arthritis (RA), multiple sclerosis (MS), systemic lupus erythematosus (SLE), and T1D (Table 1) (30, 31). The PTM autoantigens illustrated in Tables 1 and 2 represent many of the notable modified self-proteins that trigger autoreactive B cell and T cell responses and, in many cases, are specific diagnostic biomarkers as well as a reflection of disease pathology. Other PTMs (Figs. 1 and 2) can be directly affected by tissue ROS and/or inflammatory microenvironments (carbonylation, methylation, isoaspartylation, deamidation), or be influenced by more indirect downstream pathways affected by ROS (acetylation, glycosylation, phosphorylation, citrullination).
Table 1.
Post-translational Protein Modifications Associated with Autoimmune Disease
| Modification | Disease | Antigen | References |
|---|---|---|---|
| Phosphorylation | EAE/MS | αB-crystallin | (131) |
| SLE | nucleophosmin | (67) | |
| snRNP | (98) | ||
| Glycosylation | CIA | Type II collagen | (33) |
| Citrullination (Deimination) | EAE/MS | MBP | (146) |
| CapZα1 | (89) | ||
| Histone H4 | (95) | ||
| Fibrin | (88) | ||
| Type I, II collagen | (126) | ||
| α-enolase | (61) | ||
| T1D | GRP78 | (116) | |
| GAD65 | (91) | ||
| Acetylation | EAE | MBP Ac1-11 | (151) |
| SLE | Histone H2B | (75) | |
| RA | Vimentin | (57) | |
| Hydroxylation | CIA | Type II collagen | (6) |
| Methylation | SLE | Sm D1, D3 | (12, 15) |
| Deamidation | Celiac disease | Gliadin | (66) |
| T1D | Preproinsulin/Proinsulin | (93, 130) | |
| IsoAsp formation | SLE | snRNP D | (82) |
| Histone H2B | (27) | ||
| Oxidation | SLE | oxLDL | (45) |
| T1D | Insulin | (83) | |
| Carbamylation | RA | A1T1 | (134) |
| Vimentin | (107) | ||
| GRP78 | (149) | ||
| Carbonylation | T1D | P4Hb | (Unpublished data) |
A1T1, alpha 1 anti-trypsin; CapZα1, F-actin capping protein alpha-1 subunit; CIA, collagen-induced arthritis; EAE, experimental autoimmune encephalomyelitis; GAD65, glutamic acid decarboxylase 65; GRP78, glucose-regulated protein 78; LDL, low-density lipoproteins; MBP, myelin basic protein; MS, multiple sclerosis; P4Hb, prolyl-4-hydroxylase beta; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; T1D, type 1 diabetes.
Table 2.
Post-translational Protein Modifications in Type 1 Diabetes
| Target proteins | Modification | References |
|---|---|---|
| GAD65 | Citrullination | (4, 92) |
| Preproinsulin/proinsulin | Deamidation | (93, 130) |
| Insulin | Oxidation | (83) |
| IA-2 | Citrullination | (92) |
| Deamidation | (130) | |
| GRP78 | Citrullination | (4, 116) |
| ZnT8 | Citrullination | (92) |
| Deamidation | (130) | |
| IAPP | Citrullination | (4) |
| IGRP | Citrullination | (92) |
| Deamidation | (130) | |
| ICA69 | Deamidation | (130) |
| SERCA2a | Carbonylation | (122) |
| P4Hb | Carbonylation | (Unpublished data) |
IA-2, islet antigen-2; IAPP, islet amyloid polypeptide; ICA69, islet cell autoantigen 69; IGRP, islet-specific glucose-6-phosphatase catalytic subunit-related protein; SERCA2a, sarco/endoplasmic reticulum Ca2+ATPase; ZnT8, zinc transporter 8.
FIG. 1.
Structures of common post-translational protein modifications in autoimmune disease. aThe majority of these modifications are mediated by specific enzymes that for the sake of clarity are omitted from this table. GlcNac, N-acetyl-D-glucosamine; GalNac, N-acetyl-D-galactosamine; Gal, D-galactose; Nan, N-acetyl-neruaminic acid.
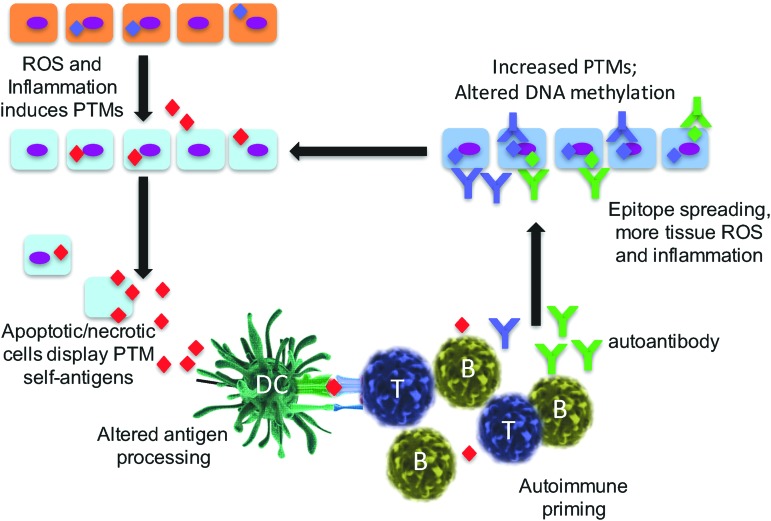
FIG. 2.
ROS and inflammation initiate cycles of autoimmunity and epitope spreading. Post-translationally modified self proteins arise in tissues during cellular stress, including ROS, inflammatory cytokines, and/or infection. PTMs are released into the milieu and phagocytized by APCs (either macrophages, dendritic cells, or B cells). Neoantigenic PTM self-peptides are then presented to autoreactive T and B cells that have escaped negative selection in the thymus and bone marrow. This occurs because the modified peptide is typically not presented during selection in noninflamed secondary lymphoid organs. Subsequently, autoreactive T and B cells infiltrate host tissue where an autoimmune response develops, leading to a second round of PTM generation and/or altered DNA methylation. APC, antigen-presenting cell; PTM, post-translational modification; ROS, reactive oxygen species. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Citrullination
One well-studied illustration of autoimmunity to a post-translationally modified self-antigen are the autoantibodies arising to citrulline-modified proteins, notably in RA. Citrulline is a product of peptidylarginine deiminase (PAD) activity on arginine residues within peptides (8). More importantly, in the case of RA, PADs (PAD1, PAD2, PAD3, PAD4) are activated by Ca2+ ion deposition within an inflamed joint (78, 142) provoking synthesis of elevated levels of citrullinated self-proteins. Sera from patients with RA recognize many citrullinated autoantigens, including collagen, vimentin, fillagrin, and α-enolase (62, 141). The early onset period of RA is marked by anti-citrulline autoantibodies that reflect disease severity (62, 141). Anti-citrulline autoantibody titers are now routinely used for the diagnosis of RA (142). In particular, circulating citrulline, arising from arginine modification, is a byproduct in the synthesis of nitric oxide, itself, a product of reactive nitrogen species (41, 85, 150). As detailed herein (Fig. 1 and Tables 1 and 2) and in accompanying monographs of this issue, B and T cell responses directed at PTMs have become important diagnostic tools for many autoimmune diseases, including an emerging group of PTM biomarkers that are important in T1D (Table 2). A recent review from Nguyen and James (105) has carefully defined the biological implications of citrulline modifications and autoimmunity arising from pancreatic beta cell proteins in the development of T1D. In particular, both citrullinated glutamic acid decarboxylase 65 (GAD65) and glucose-regulated protein 78 (GRP78) elicit a vigorous B and T cell autoimmune response in human T1D and nonobese diabetic (NOD) mouse murine disease, respectively (92, 116). The latter studies were marked by a dramatic and aberrant upregulation of PADI2 in the islets of NOD mice, supported by genetic risk in the Idd25 locus of mouse chromosome 4. These citrulline modifications of T1D autoantigens were linked to cytokine and/or ROS stress effects on the endoplasmic reticulum, a recurrent theme in fostering many PTMs.
Carbonyl PTMs
Protein carbonylation is a major product of tissue proteins in response to oxidative stress. Increased carbonylation of self-proteins arises due to decreases in antioxidant defense pathways, increases in ROS production, or an inability to remove or repair oxidized self-proteins, as previously reviewed by Nystrom (106). The classical antioxidant (ROS) defense mechanisms, superoxide dismutase, catalases, and peroxidases, all protect against the induction of carbonyl modifications. Carbonylation is a metal-catalyzed (free iron) oxidative modification of the side chains of proline, arginine threonine, and lysine (Fig. 1). Carbonyl modifications are typically more difficult to induce relative to other oxidative modifications. This modification has many deleterious effects on various intracellular enzymatic mechanisms, and mitochondria are particularly vulnerable to ROS-induced carbonylation (106). Carbonylation is a marker of cellular senescence and is increased in aging cells and tissues. There is evidence to suggest that carbonylation is one protective mechanism of cells for directing damaged proteins into proteolytic degradation pathways, as these modified proteins are conformationally unstable. Carbonylation is an irreversible modification; thus, the biological functions of these modified proteins are unrepairable (32). All of these properties make carbonylated self-proteins recognized in an autoimmune response, particularly if they fail to find their way to normal cellular degradation pathways.
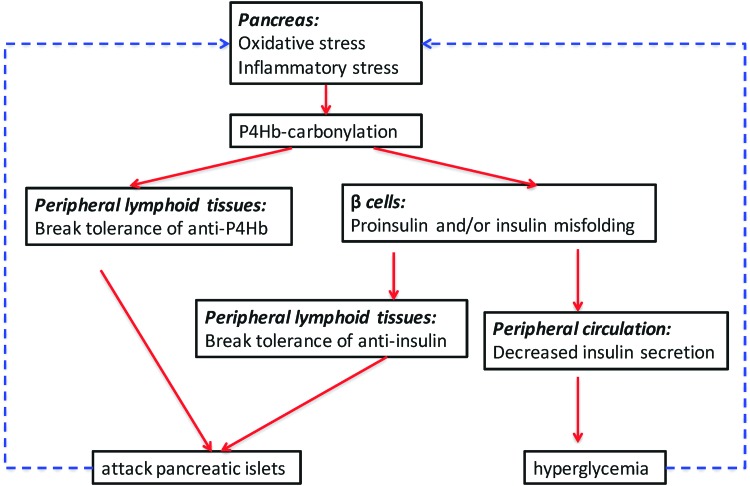
It has been observed that increases in ROS contribute to insulin resistance and metabolic dysfunctions in adipose tissue of both animal models and human type 2 diabetes (T2D) (36, 119, 124). Although publications have profiled carbonylated plasma proteins as potential biomarkers in T2D (35, 42), little is known about carbonylation in the progression of T1D. Our recently submitted studies from Yang et al. defined a group of pancreatic beta cell proteins with carbonyl PTMs, all bound by autoantibodies from human and NOD mice T1D antisera. Among this group were both novel and established biomarkers of T1D, including protein disulfide isomerase (PDI) isoforms, 14-3-3 protein isoforms, GRP78, chymotrypsinogen B, and malate dehydrogenase. Of interest, carbonylated prolyl-4-hydroxylase beta (P4Hb, also known as protein disulfide isomerase A1; PDIA1) was found to be an early autoantigen in both human and murine models of T1D. P4Hb is essential for the appropriate folding of insulin from pancreatic beta cells (112). Our data suggest a novel role of modified P4Hb, both as an early target of autoimmunity and as a pathway that provokes autoimmunity to insulin and/or proinsulin. In fact, autoimmunity to P4Hb always preceded autoimmunity to insulin in both NOD mice and human T1D, as defined by the pathway described in Figure 3. Carbonylation is amplified by either oxidative or cytokine stress to beta cells. Carbonylated P4Hb fails in its ability to accurately fold and process proinsulin to insulin. We hypothesize that misfolded proinsulin levels accumulate in the cell or serum and lead to linked autoimmune responses to insulin itself. Aberrant carbonylated P4Hb biological functions in the beta cell provide an explanation for recent observations of increased proinsulin to insulin ratios in the progression of T1D (115).
FIG. 3.
ROS and inflammation induce carbonyl modification of the chaperone protein, P4Hb. The oxidative and cytokine stress in the pancreatic islet microenvironment induces carbonyl modification of beta cell proteins (see text) and P4Hb. P4Hb is one chaperone protein that is responsible for the accurate folding and processing of proinsulin to insulin in the beta cell. Carbonyl-modified P4Hb is a neoantigen that induces autoreactive B and T cells found in early onset human T1D and in the NOD mouse. In addition, carbonyl P4Hb fails to accurately process proinsulin, leading to reduced insulin secretion. NOD, nonobese diabetic; P4Hb, prolyl 4-hydroxylase beta polypeptide; T1D, type 1 diabetes. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
ROS and PTMs in the development of NETs
NETosis is a neutrophil destruction mechanism differing from the apoptotic pathway, initiated when neutrophils confront microorganisms, and generating chromatin webs from intracellular contents (22). The extruded DNA webs carry a number of bound bactericidal proteins (lactoferrin, elastase, proteinase 3, myeloperoxidase, cathepsin G, etc.) as well as histones and granule proteins. This pathway is marked by the release of mitochondrial DNA, a process dependent on cellular stress created by ROS, superoxide anion, hydrogen peroxide, myeloperoxidase, and NADPH oxidase. Patients who are deficient in myeloperoxidase cannot generate NETs. The mitochondrial electron transport chain is one source of intracellular ROS in neutrophils. With relevance to autoimmune responses, NETs are believed to be one source of immunogenic histone H2B. A study by Liu et al. (75) identified the PTMs originating from the histones of NETs. Autoantibodies were elicited to citrullinated forms of histone H3 and H4 and to forms of histone H2B that were either methylated or acetylated. The two pathways of cellular destruction, either apoptosis or NETosis, trigger unique PTMs, all of which may break immune tolerance to modified self-proteins. The role of NETs in T1D has yet to be fully resolved; one recent study illustrated a reduction in serum components of NETs (proteinase 3 and neutrophil elastase), consistent with a reduced overall neutrophil count in early onset T1D (111). Conflicting studies report increases in these same NET components in T1D (139).
A large number of PTM cytoplasmic proteins are the autoantigenic targets of autoantibodies in SLE (Table 1). Tissue pathology, inflammatory cytokines, and ROS create intracellular metabolic stresses that favor the generation of PTMs, including carbonyl modifications of self-proteins as detailed herein (31, 32). Pathways designed to clear inflammatory material (NETosis and apoptosis) may drive the PTM of self-antigens, being viewed as “foreign” to the immune system and are amplified by ROS. Although ROS are important in protecting cells from infectious agents (such as bacteria), ROS also induce aberrant PTMs within self-proteins that may be immunogenic in the host. The abnormal clearance of apoptotic or NETotic cellular debris is believed to be a principal source of autoantigens in lupus (46, 47, 65, 127). As noted earlier, NETs are enriched in various PTMs of self-proteins, including citrullination (58). In addition, specific lupus autoantigens, including nucleosome dsDNA, snRNPs, Ro/SSA, and La/SSB, are found within cell surface blebs in apoptosis (13), a reservoir of modified proteins. In addition, PTMs such as transglutamination, phosphorylation, proteolysis, and adenosine diphosphate-ribosylation are amplified in apoptosis (129). Notably, phosphorylated SR proteins (also known as pre-mRNA splicing factors) are targets of human SLE autoimmunity (104).
Isoaspartyl modification, the consequence of spontaneous, nonenzymatic isomerization of aspartic acid, is amplified in the presence of ROS as well as in cells that undergo necrosis and/or apoptosis (Fig. 1 and details below) (16, 19). Both lupus-prone MRL mice and human SLE patients have elevated titers of autoantibodies that react with isoAsp-modified histone H2B (27). Similarly, symmetric dimethyl arginines found in the C-terminus of the snRNP complex are amplified by ROS (12).
PTMs can trigger more extensive autoimmunity, such as epitope spreading
We and others have identified significant differences between the presence and specificity of T cell and B cell immunity that arise in response to ROS stress-induced PTM self-proteins (27–31, 39, 45, 82, 87, 92). In particular, T cell responses to PTM determinants are most often specific for the modified peptide form only and do not bind the native (unmodified) protein or peptide. This concept is demonstrated in mice immunized with the isoAsp snRNP D where T lymphocytes bind only to the isoAsp-modified snRNP D peptide and are unresponsive to the native aspartic acid form of peptide (82). T cell autoimmunity to PTM self-proteins will be more fully addressed by other authors in this issue of Antioxidants and Redox Signaling. In contrast, B lymphocyte and autoantibody responses are typically promiscuous in their binding to both the modified and native self-protein. However, tolerance is often not only broken to the PTM-modified self-protein but also exhibits cross-binding to the native protein. This phenomenon may be due to the ability of antibodies to bind amino acid sequences adjacent to the PTM that is present in both modified and native protein sequences. As one example, human SLE and lupus-prone MRL/lpr mice have autoantibodies that are cross-reactive to both isoAsp and aspartic acid forms of histone H2B (27). It was found that autoimmunity is initiated with the PTM self-protein/peptide, and thereafter spreads in an intra- and extra-molecular manner to other determinants. Thus, the presence of a PTM in a self-protein amplifies “epitope spreading,” a mechanism in which the immunity amplifies to include determinants outside the site(s) that trigger the original immune response (Fig. 2) (70). As a consequence, there is both intra- and inter-molecular B and T cell epitope spreading to self-antigens in T1D, SLE, and MS (59, 82). Epitope spreading is associated with the development, progression, and severity of autoimmune disease (3). One favored mechanism is that modified self-antigens are recognized as foreign, thus triggering a restricted autoimmune response, even before the appearance of symptoms, followed by the subsequent accumulation of additional targeted determinants (Fig. 2). As noted earlier, necrotic and apoptotic cells are early sources of altered self-proteins and amplified by tissue stress, notably stress from ROS (38) or altered pH (39).
PTMs in antigen processing and presentation
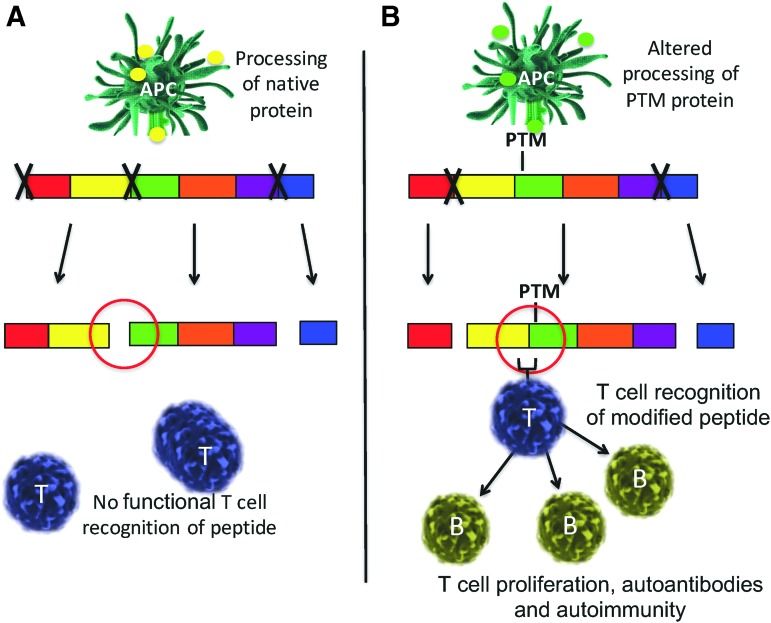
Autoimmune responses develop to modified self-proteins due to defects in the negative selection of immune cells. It is clear that accurate antigen processing plays a major role in peptides that control negative selection (84). However, the presence of a PTM residue that is critical for cleavage changes the specificity of proteases and the types and sequences of self-peptides generated and, ultimately, presented to the immune system (Fig. 4). The presence of specific PTMs within a processed peptide also affects the binding to major histocompatibility complex (MHC). As one example, the absence of N-glycosylation of glutamate receptor subunit 3 in Rasmussen's encephalitis reveals sites that are susceptible to granzyme B cleavage, thereby creating a novel autoantigen (neoepitope) (37). Many proteases fail to recognize the β-peptide isoform between isoAsp residues and the adjacent carboxyl side amino acids (56). In addition, studies illustrated that proteins are resistant to asparagine endopeptidase cleavage as a result of the spontaneous deamidation of asparagine residues (100). Simply put, the proteolytic enzyme recognition of PTM self-proteins may generate novel repertoires of peptides during antigen presentation and the establishment of immune tolerance (Fig. 4). These observations were confirmed several years ago in studies of model proteins in immunity (79–82), now confirmed by more recent work with disease-relevant PTM autoantigens. Cathepsin D, an enzyme in the antigen processing pathway, differentially cleaves substrate proteins based on the presence or absence of isoaspartyl protein modification (29). This phenomenon is also supported by the cleavage products of Granzyme B, again altered by the presence of post-translationally modified self-protein substrates, leading to neo-epitope presentation (14).
FIG. 4.
Antigen processing is altered by PTMs. (A) Native isoforms of self-antigens are cleaved by intracellular proteases (as represented by “X”) into distinct peptides. Under most conditions, negative selection eliminates T and B cells that recognize these normal isoform peptides due to clonal deletion and anergy. (B) Post-translationally modified sites are often not accurately recognized or cleaved by proteases, thereby creating novel self-peptides to which immune tolerance does not exist. Novel peptide presentation by APCs primes T cells, which provide help to B cells in secreting autoantibodies. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The biochemical processing within acidified lysosomal compartments of antigen-presenting cells (APCs) dictates the types of short peptides subsequently presented on both class I and class II MHC. Interesting studies by Ireland et al. (54) found that citrulline-modified peptides were generated, processed, and presented to CD4 T cells only in B cells that undergo autophagy. The presentation of unmodified peptides was unchanged. Thus, it was the autophagy pathway itself that presumably created the citrulline PTM that was subsequently presented in the priming of CD4 T cells. Our laboratory and others have demonstrated the unique ability of B lymphocytes as APCs in presenting specific self-peptides to T cells. For example, B cells transfer processed antigens to other APCs, including macrophages and dendritic cells (43, 44, 113). Thus, different APC subsets themselves may control the determinants generated and eventually presented by the immune system (20, 100).
Algorithms that predict motifs of how particular peptides will associate with MHC do not incorporate the presence of peptide PTMs. The “fit” of the PTM peptide for MHC differs significantly from native (unmodified) peptide. The MHC binding of some post-translationally modified T1D autoantigens has been studied and reviewed by McGinty et al. (91, 92). Also, different modifications of myelin basic protein (MBP) change the affinity of peptides for MHC compared with the corresponding wild-type peptide (21). Acetylated MBP peptide (Ac 1–11) stimulates pathogenic T cells in murine MS, though the unmodified peptide binds MHC with virtually identical kinetics. Similarly, isoaspartic acid residues in cytochrome c or snRNP D peptides bind MHC class II in a manner that is identical to the unmodified peptides (82), yet immune tolerance is maintained only to the native peptide. Alternatively, citrullinated-modified peptides of RA autoantigens, specifically vimentin, bind the high-risk allele, human leukocyte antigen (HLA)-DRB1*0401 greater than the native (unmodified) peptide (49). The overall message from these collective studies is that PTM self-peptides may or may not be processed and bound by MHC in a manner found with unmodified (native) peptide. Moreover, it cannot be predicted whether T cell receptors (TCRs) and B cell receptors will bind PTM self-peptides or proteins and cross-react with the corresponding unmodified antigen.
ROS alter the methylation of DNA and protein
Another emerging PTM of significance is intracellular methylation modification, a process now known to be essential in the normal functions of all immune cells. The enzymatic addition of methyl groups to substrates (including proteins, lipids, and DNA) occurs via the single methyl donor in cells, S-adenosyl-methionine (SAM). The family of methyltransferases includes both protein methyltransferases (PRMTs) and DNA methyltransferases (DNMTs). ROS alter all of these methylation pathways (10, 99, 145). For example, oxidation of DNA is required for DNA methylation pathways by ten-eleven translocations (TETs) (reviewed in Ref. 10).
The most well-studied transmethylation substrates in regulating epigenetics are histone proteins and, of course, DNA. However, other substrates of protein methylation are key regulators in a diverse array of cellular pathways, including lymphocyte signaling and differentiation pathways and the regulation of transcription. In fact, lymphocytes require accurate protein methylation during cellular activation compared with most other cell types (Table 3) (40). Protein methylation is required for efficient TCR signaling, and in the regulation of cytokine responses (including interferon [IFN]α/β-induced transcription via STAT1; Table 3). T cell growth and development relies on the accuracy of histone methylation and promotor region CpG dinucleotide sequences. Inhibition of protein arginine methyltransferase (PRMT) alters the ability of phosphorylated signal transducers and activators of transcription (STAT) in binding to DNA (103). Moreover, PRMT activity and Vav1 methylation is increased in T cells on CD28 costimulation (9). The recurring themes of this review are the effects of ROS in these various, and sometimes indirect, biological pathways. Recent studies have demonstrated that PRMT1, a major protein methyltransferase in humans, is downregulated by ROS in the microenvironment, causing the release of asymmetric dimethylarginine into the serum, a biomarker of tissue pathology (99).
Table 3.
PRMTs Associated with Immune Responses
| Methylation product | PRMTs member | Epigenetic regulation | References | |
|---|---|---|---|---|
| Type I | Methylation in the terminal guanidine nitrogen atoms Monomethylation Asymmetric dimethylation |
1 (RMT1 in yeast) 2 3 (RMT3 in yeast) 4 (CARM1) 6 8 |
Th2 cytokine production (IL-2 and IL-4) (PRMT1) Signal transduction in T cells Vav1, STAT1/6, AKt/PKB, NIP45, and NFAT (PRMT1) STAT3 (PRMT2) Interaction to Interferon receptor (PRMT1) P13K kinase pathway in B cell differentiation (PRMT1) T helper cells function (PRMT2) Thymocyte differentiation (PRMT4) Th17 differentiation (PRMT2, 4, and 6) Histone methylation, H4R3 (PRMT1) and H3R2, R8, R17, and R26 (PRMT4) |
(63, 125, 143) (68, 102, 103, 120) (140) (53) (55) (60) (64) (7, 144) |
| Type II | Methylation in the terminal guanidine nitrogen atoms Monomethylation Symmetric dimethylation |
2 5 9 (FBXO11) Hsl7 |
IL-2 gene expression (PRMT5) Histone methylation, H2A, H4 (PRMT5) B cell lymphoid cancer (PRMT5) |
(114) (34, 108) (138) |
| Type III | Methylation in the terminal guanidine nitrogen atoms Monomethylation |
7 | Histone methylation, H4 (PRMT7) Thymus and dendritic cells (PRMT7) |
(96) (69, 97) |
| Type IV | Methylation in the internal guanidine nitrogen atoms Monomethylation |
RMT2 (in the yeast) |
CARM1, coactivator-associated arginine methyltransferase 1; NFAT, nuclear factor of activated T cells; PRMT, protein arginine methyltransferase.
Autoimmune disease is marked by abnormal methylation modifications, both protein and DNA. Defects in the methylation status of CpG dinucleotide elicit T cell autoimmunity, contributing to the pathogenesis of SLE (123). As mentioned earlier, symmetric dimethylated ribonucleoproteins, snRNP proteins D1 and D3, are autoantigens recognized by autoantibodies in SLE patients (12). Similarly, arginine-methylated MBP triggers the autoimmunity, in both the B and T cell compartments, in MS (110).
In T1D, it is attractive to hypothesize that DNA and/or protein methylation pathways are influenced by ROS as they are by inflammatory cytokines (IL-1β, IFNγ, or IL-6) in the pancreatic islet microenvironment (118). From the time that autoantibodies are first detected in individuals at genetic risk for T1D, the course of disease is highly variable (132). Many individuals do not progress to overt disease, and those who do may progress over different periods ranging from a few months to decades. This variability has largely focused on the immune effector and regulatory cells that are involved in β cell killing and maintenance of tolerance (11, 90). However, it is equally likely that β cells respond to the immune attack and environmental factors such as ROS in ways that may accelerate or retard disease progression (Fig. 5). In addition to the modification of these proteins, there may also be modifications of the enzymes responsible for the epigenetic changes themselves. TET2 was reported to be modified by acetylation during oxidative stress, and DNMT3a has been reported to be SUMOylated, which modulates its repression of transcription (74, 152). DNMT3a is known to control histone deacetylase (HDAC) gene expression and in the setting of inflammation, TET2 physically associates with certain HDACs (23). TET2 is required to resolve inflammation due to IL6 (72).
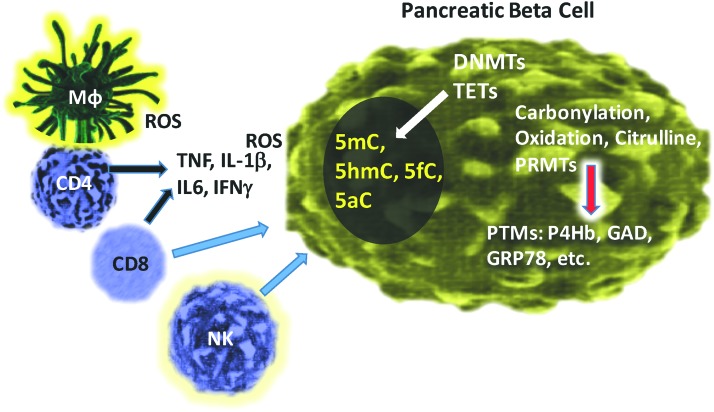
FIG. 5.
The generation of PTMs in pancreatic beta cells in T1D. Immune cell infiltration of pancreatic islets includes macrophages, CD4 and CD8 T cells, and NK cells. Direct attack to the beta cell from CD8 and NK cells occurs, whereas ROS is released from resident macrophages. Other inflammatory cytokines, including TNFα, IL-1β, IL6, and IFNγ, all contribute to PTMs generated inside of the beta cell. The response to ROS and cytokine stress includes various PTMs such as carbonylation, oxidation, citrullination, and protein methylation. In addition, DNMTs and TETs altered by the presence of ROS cause various defects in DNA methylation and subsequent downstream translational regulation. DNMT, DNA methyltransferase; IFN, interferon; TET, ten-eleven translocation. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Rui et al. have studied individuals who were at very high risk for diabetes by virtue of finding two or more positive autoantibodies and dysglycemia (117, 118). Historically, ∼75% of these subjects progress to overt diabetes within 5 years. These studies measured the relative levels of unmethylated insulin INS DNA (released from dying β cells) in the serum compared with the amount of methylated INS DNA. It was found that the frequency of increased levels of unmethylated INS DNA measurements (taken about every 6 months) was low in the individuals at risk, and not significantly different in progressors and nonprogressors to diabetes. However, the frequency of elevated levels of unmethylated INS DNA was significantly higher in the very high-risk subjects, suggesting increased β cell killing in the prediagnosis period. These observations, and others from clinical studies of β cell function and pathology, have refined the kinetics of T1D progression, as originally reported by Eisenbarth in 1986.
In summary, studies of β cells during the progression of T1D in NOD mice and human β cells in vitro indicate that there is induction of DNMTs as well as PTMs that can remodel or affect survival of β cells (117, 118). An underlying theme to these studies is that the processes of epigenetic protein modifications are interconnected, possibly both in response to inflammatory mediators or more directly in which one process modifies the others. An overview of the converging pathways is illustrated in Figure 5.
Protein arginine methyltransferase
The first PRMT enzyme was identified, cloned, and characterized in 1996 (73). In the past 20 years, an additional 11 PRMT enzymes, with varying biological substrate activity, have been discovered, but all still utilizing SAM as the sole intracellular methyl donor. All of the PRMTs will methylate in a unique sequence pattern, based around glycine-arginine-rich domains. Four classes of PRMTs exist, depending on the unique methylation products that are catalyzed (Table 3). With relevance to autoimmunity, type I PRMTs catalyze asymmetric di-methylarginine modification and type II PRMTs will form symmetric di-methylarginine, both of which are known antigenic targets of lupus autoimmunity, notably the snRNP D protein (7). Other PRMT family proteins, described later, control many aspects of immune cell differentiation and function (Table 3).
The first well-studied methyltransferase, PRMT1, catalyzes more than 85% of arginine methylation within cells (7). PRMT1 regulates the production of T cell cytokines, both IL-2 and IL-4, via the methylation of Vav1, STAT1/6, and Akt/PKB in the T cell signaling pathway (9, 94, 103, 109) and by the regulation of transcription via nuclear factor of activated T cells (NFAT) (102). B lymphocyte differentiation is regulated by PI3K kinase activity that is optimized by PRMT1-mediated methylation. It is now clear that PRMT1-directed methylation of histones H4R2, H3, and H4 mediates DNA transcriptional activation (51, 71, 137). Yet other histone PTMs, including acetylation, in T lymphocytes are linked with disease flares in human SLE (50) whereas Th17 cell differentiation is regulated by PRMT2-mediated methylation (133). Reduced cellular methylation due to the loss of PRMT4 (coactivator-associated arginine methyltransferase 1 [CARM1]) alters normal thymocyte differentiation, chromatin remodeling, and possibly tolerance to self proteins (2). Finally, alterations in the SAM- and/or PRMT-mediated methylation pathways influence IL-2, IL-4, and IFNγ gene expression from T lymphocytes.
As detailed herein, ROS-mediated PTM within T cells alters the production of inflammatory cytokines that contributes to the onset of autoimmunity. Human SLE is marked by aberrant TCR signaling that is caused, in part, by an increase in phosphorylation of ERK, Syk, ZAP-70, and PI3K kinase activity (18, 101). As illustrated here, ROS may initiate a chain reaction of downstream effects of methylation and phosphorylation leading to aberrant T cell biology. The overall implications are that ROS-mediated alterations in PRMT methylation of histones contribute to increases in the serum levels of T lymphocyte products, including IFNγ, IL-4, and IL-17, inflammatory mediators in human SLE (1, 128) (Table 3).
Repair of PTMs induced by ROS
Isoaspartyl PTMs are created by a number of cellular stresses, including ROS. The efficient repair of cellular isoaspartyl PTMs is by the enzyme known as protein l-isoaspartyl (d-aspartyl) methyltransferase (PIMT). The inability to repair isoaspartyl PTMs in lymphocytes leads to lupus-like autoimmunity in animal models (28, 147). The PIMT repair pathway is essentially a response to ROS and inflammation in tissues, in attempts to repair deleterious isoaspartyl PTMs to the native (aspartic acid) form in self-proteins. PIMT knockout animal models, or a failure to repair the isoaspartyl PTM, causes a five-fold increase of isoAsp proteins in both lymphoid cells (B and T cells), and in all somatic cells, leading to an autoimmune phenotype and premature death (28, 76). The highly conserved PIMT enzyme is found in both prokaryotes and eukaryotes, thus emphasizing its importance in maintaining cellular health. Isoaspartyl PTMs are found in several autoantigens in SLE, including histone H2B and snRNPs (82, 148). The inability to repair cellular isoaspartyl self-proteins is believed to trigger pathologic autoimmunity in a manner similar to other PTM self. This is but one of the indirect, or downstream consequences of ROS stress on tissues.
In the murine model of human SLE, the MRL mouse experiences increased isoaspartyl PTM proteins in the kidney and brain, two sites of immune complex-mediated pathology (147). Increased isoaspartyl protein levels lead to T cell hyperproliferation, a phenotype of both human and murine SLE (28). We have recently identified the presence of isoAsp PTMs within several residues of the T cell signaling protein, ZAP70 (unpublished data), likely contributing to the aberrant T cell hyperproliferative phenotype in SLE.
Regarding T1D, the PIMT isoaspartyl repair enzyme is highly expressed in pancreatic beta cells and in the transformed insulinoma cell line, INS-1. Interestingly, the induction of PIMT expression delays the appearance of symptoms and reduces the severity of T1D in the BB rat model of T1D (135). The PIMT1 gene maps to a region, 6q24–25, linked to the IDDM5 site studied in genome analyses of human T1D (136). Four PIMT1 polymorphisms were found to be in linkage disequilibrium, with PCMT1 promotor activity increased in response to cytokine stimulation. Collectively, the work supports an interaction between PCMT1 and both HLA and SUM04 in the genetic risk for T1D. As yet, however, specific PIMT polymorphisms already defined to exist in humans (17, 25, 26) have not yet been clearly associated with any autoimmune syndrome. Moreover, the expression of PIMT protects from Bax-induced cellular apoptosis, perhaps yet another mechanism that evolved to prevent the release of isoaspartyl-modified self-proteins (52). We have recently observed significant increases in cellular isoaspartyl levels in the islets of NOD diabetogenic mice as well as in human pancreatic islets treated with physiologic levels of H2O2 or inflammatory cytokines (Yang and Mamula, unpublished data). The emerging picture is that ROS and/or cytokine-induced inflammation of tissues trigger various PTM pathways, followed by protection mechanisms initiated by the cell to both prevent its destruction and repair aberrant self-proteins. Unfortunately, the inflammatory storm found in autoimmune disease is often too vigorous to be impeded by these protective intracellular mechanisms. Further studies will be required to fully appreciate both the direct effects of ROS and more indirect pathways altered by ROS in T1D.
ROS, DNA methylation, and epigenetics in autoimmunity
Epigenetics refers to gene expression that is influenced and/or controlled by various modifications that arise during the transcription and translation of genetic loci (77). These epigenetic modifications collectively include DNA and histone methylation, histone acetylation/deacetylation, and nucleosome remodeling. As detailed earlier, post-translationally modified proteins, many of which are altered by ROS, are linked to alterations in DNA methylation. One study revealed that overall hypomethylation of DNA, in particular, methyl cytosine residues, is more frequent in SLE as compared with healthy individuals (5).
Relevant to human T1D, Rui et al. reported epigenetic modifications of β cells during progression of diabetes in NOD mice, the murine model of T1D (118). The study demonstrated specific methylation of exons in Ins1 and the promoter and exons of the Ins2 gene. More careful analyses revealed an inverse relationship between methylation states of Ins2 Exon1 and Ins2 mRNA levels in β cells. The study observed an induction of DNMT3a during disease progression, which was shown to account for the epigenetic changes by siRNA silencing. The progression of islet pathology was coincident with the induction of DNMT3a and the methylation of Ins2 DNA in response to inflammatory cytokines (IL-1β, IL-6, and IFNγ). DNMTs are critically involved in β cell differentiation, and subsequent studies suggested that there may be cellular changes resulting from the inflammation (cytokines or ROS) that may result from epigenetic modifications (117). A subpopulation of dedifferentiated β cells in the islets of NOD mice develop during progression of disease, noted by the loss of normal differentiation features of β cells and the acquisition of stem-like characteristics. The novel β cell subpopulation showed increased frequency of methylation of CpG sites in the Ins genes compared with normal β cells (unpublished). These β cells were resistant to immunologic killing, suggesting that the mechanism of epigenetic modification to inflammatory mediators may represent a cellular protective response.
In support of the studies in T1D described earlier, hypomethylation of DNA is a biological feature and profiling characteristic of a number of autoimmune maladies, including MS, Grave's disease, mixed connective tissue disease, ankylosing spondylitis Addison's disease, cancer, and Alzheimer's disease (48).
Concluding Remarks
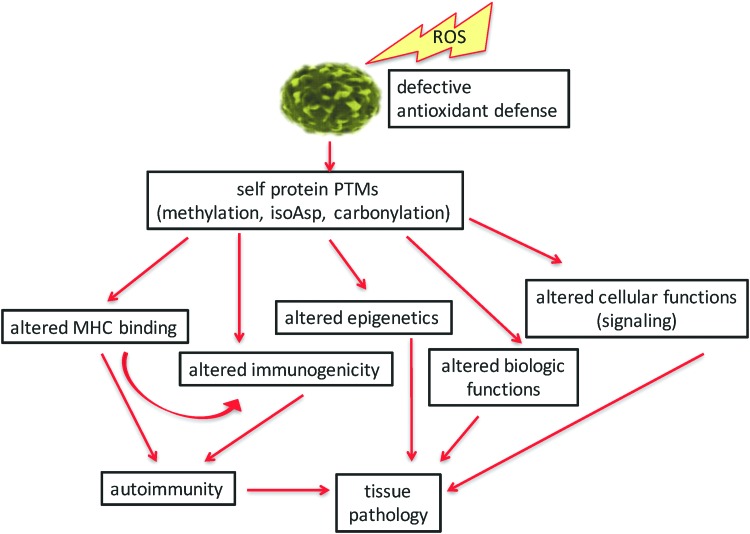
We have illustrated several specific ROS-influenced pathways that influence the onset and progression of autoimmunity (Fig. 6). Many of the PTMs and antigen processing pathways described herein are spontaneous in nature and beyond the prediction of genetics. It is now obvious that many PTMs and cellular pathways are affected by ROS. Indeed, it may appear that ROS are at the “center of the universe” of triggering autoimmunity. ROS trigger several PTMs of self-proteins in direct ways, such as with carbonyl or isoaspartyl modifications. Indirect pathways are affected by ROS, as with methylation of DNA or proteins and with amplifying enzyme-mediated PTMs (citrullination) and by triggering PTM repair mechanisms (PIMT). Although we have defined some specific PTMs in T1D, SLE, and many other autoimmune syndromes, this dynamic and quickly changing field does not allow us to enumerate and detail all published PTM autoantigens. Described herein, ROS-amplified PTMs lead to alterations in self-peptide binding to MHC, defects in immune tolerance induction, altered or defective biological functions of self proteins, changes in epigenetics, and even changes in cellular metabolic pathways (such as signaling). The end product of these pathways is the development of autoimmune-mediated tissue pathology (Fig. 6). The fine specificity of B and T cell autoimmunity, such as T cell subset analyses, and pathological responses to PTM self-proteins are detailed within other manuscripts of this volume of Antioxidants and Redox Signaling. Studies by Piganelli and colleagues (reviewed in this volume) have carefully defined the role of inflammatory and oxidated stress in pancreatic tissue and the endoplasmic reticulum in the course of T1D autoimmunity (86, 87). The emerging technologies in proteomics and tissue analyses will undoubtedly change the landscape of this field in the coming months and years. These analyses have already “modified” how the clinical community diagnoses and assesses the progression of disease, and tissue pathology. With the identification of specific biomarkers and an understanding of their origins, the field will now have potential therapeutic pathways as targets to modify these autoimmune diseases.
FIG. 6.
Overall pathway of ROS leading to autoimmune pathology. The origins of autoimmunity begin with a tissue microenvironment that is rich in ROS combined with weakened or overwhelmed antioxidant defenses. Various PTMs arise in surviving cells that alter several cellular pathways, including immunity (MHC binding and altered immunogenic self proteins) and changes in epigenetics, protein functions, or cellular pathways. The collective outcomes of these changes lead to tissue pathology. MHC, major histocompatibility complex. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Abbreviations Used
- APC
antigen-presenting cell
- CARM1
coactivator-associated arginine methyltransferase 1
- DNMT
DNA methyltransferase
- GAD65
glutamic acid decarboxylase 65
- GRP78
glucose-regulated protein 78
- HDAC
histone deacetylase
- HLA
human leukocyte antigen
- IFN
interferon
- MBP
myelin basic protein
- MHC
major histocompatibility complex
- MS
multiple sclerosis
- NFAT
nuclear factor of activated T cells
- NETs
neutrophil extracellular traps
- NOD
nonobese diabetic
- P4Hb
prolyl 4-hydroxylase beta polypeptide
- PAD
peptidylarginine deiminase
- PDI
protein disulfide isomerase
- PIMT
protein l-isoaspartyl (d-aspartyl) methyltransferase
- PRMT
protein arginine methyltransferase
- PTM
post-translational modification
- RA
rheumatoid arthritis
- ROS
reactive oxygen species
- SAM
S-adenosylmethionine
- SLE
systemic lupus erythematosus
- STAT
signal transducer and activator of transcription
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TCR
T cell receptor
- TET
ten-eleven translocation
Acknowledgments
This work was supported by National Institutes of Health (AR-41032 and AI-48120 and), the Lupus Research Alliance, and the Juvenile Diabetes Research Foundation to M.J.M.
References
- 1.Akahoshi M, Nakashima H, Tanaka Y, Kohsaka T, Nagano S, Ohgami E, Arinobu Y, Yamaoka K, Niiro H, Shinozaki M, Hirakata H, Horiuchi T, Otsuka T, and Niho Y. Th1/Th2 balance of peripheral T helper cells in systemic lupus erythematosus. Arthritis Rheum 42: 1644–1648, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Akimzhanov AM, Yang XO, and Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem 282: 5969–5972, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, and Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med 349: 1526–1533, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Babon JA, DeNicola ME, Blodgett DM, Crevecoeur I, Buttrick TS, Maehr R, Bottino R, Naji A, Kaddis J, Elyaman W, James EA, Haliyur R, Brissova M, Overbergh L, Mathieu C, Delong T, Haskins K, Pugliese A, Campbell-Thompson M, Mathews C, Atkinson MA, Powers AC, Harlan DM, and Kent SC. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med 22: 1482–1487, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balada E, Ordi-Ros J, and Vilardell-Tarres M. DNA methylation and systemic lupus erythematosus. Ann N Y Acad Sci 1108: 127–136, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Batsalova T, Lindh I, Backlund J, Dzhambazov B, and Holmdahl R. Comparative analysis of collagen type II-specific immune responses during development of collagen-induced arthritis in two B10 mouse strains. Arthritis Res Ther 14: R237, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedford MT. and Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bicker KL. and Thompson PR. The protein arginine deiminases: structure, function, inhibition, and disease. Biopolymers 99: 155–163, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchet F, Cardona A, Letimier FA, Hershfield MS, and Acuto O. CD28 costimulatory signal induces protein arginine methylation in T cells. J Exp Med 202: 371–377, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochtler M, Kolano A, and Xu GL. DNA demethylation pathways: additional players and regulators. Bioessays 39: 1–13, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Bonifacio E, Scirpoli M, Kredel K, Fuchtenbusch M, and Ziegler AG. Early autoantibody responses in prediabetes are IgG1 dominated and suggest antigen-specific regulation. J Immunol 163: 525–532, 1999 [PubMed] [Google Scholar]
- 12.Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, and Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 275: 17122–17129, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Casciola-Rosen LA, Anhalt G, and Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med 179: 1317–1330, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casciola-Rosen LA, Miller DK, Anhalt GJ, and Rosen A. Specific cleavage of the 70-kDa protein component of the U1 small nuclear ribonucleoprotein is a characteristic biochemical feature of apoptotic cell death. J Biol Chem 269: 30757–30760, 1994 [PubMed] [Google Scholar]
- 15.Chang HH, Hu HH, Lee YJ, Wei HM, Fan-June MC, Hsu TC, Tsay GJ, and Li C. Proteomic analyses and identification of arginine methylated proteins differentially recognized by autosera from anti-Sm positive SLE patients. J Biomed Sci 20: 27, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimmino A, Capasso R, Muller F, Sambri I, Masella L, Raimo M, De Bonis ML, D'Angelo S, Zappia V, Galletti P, and Ingrosso D. Protein isoaspartate methyltransferase prevents apoptosis induced by oxidative stress in endothelial cells: role of Bcl-Xl deamidation and methylation. PLoS One 3: e3258, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Clarke S. Perspectives on the biological function and enzymology of protein methylation reactions in eucaryotic and procaryotic cells. Adv Exp Med Biol 231: 213, 1988 [DOI] [PubMed] [Google Scholar]
- 18.Crispin JC, Kyttaris VC, Terhorst C, and Tsokos GC. T cells as therapeutic targets in SLE. Nat Rev Rheumatol 6: 317–325, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Angelo S, Ingrosso D, Migliardi V, Sorrentino A, Donnarumma G, Baroni A, Masella L, Tufano MA, Zappia M, and Galletti P. Hydroxytyrosol, a natural antioxidant from olive oil, prevents protein damage induced by long-wave ultraviolet radiation in melanoma cells. Free Radic Biol Med 38: 908–919, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Davidson HW. and Watts C. Epitope-directed processing of specific antigen by B lymphocytes. J Cell Biol 109: 85–92, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Haan EC, Wagenaar-Hilbers JP, Liskamp RM, Moret EE, and Wauben MH. Limited plasticity in T cell recognition of modified T cell receptor contact residues in MHC class II bound peptides. Mol Immunol 42: 355–364, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Delgado-Rizo V, Martinez-Guzman MA, Iniguez-Gutierrez L, Garcia-Orozco A, Alvarado-Navarro A, and Fafutis-Morris M. Neutrophil extracellular traps and its implications in inflammation: an overview. Front Immunol 8: 81, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deplus R, Blanchon L, Rajavelu A, Boukaba A, Defrance M, Luciani J, Rothe F, Dedeurwaerder S, Denis H, Brinkman AB, Simmer F, Muller F, Bertin B, Berdasco M, Putmans P, Calonne E, Litchfield DW, de Launoit Y, Jurkowski TP, Stunnenberg HG, Bock C, Sotiriou C, Fraga MF, Esteller M, Jeltsch A, and Fuks F. Regulation of DNA methylation patterns by CK2-mediated phosphorylation of Dnmt3a. Cell Rep 8: 743–753, 2014 [DOI] [PubMed] [Google Scholar]
- 24.Derbinski J, Schulte A, Kyewski B, and Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol 2: 1032–1039, 2001 [DOI] [PubMed] [Google Scholar]
- 25.DeVry CG. and Clarke S. Polymorphic forms of the protein L-isoaspartate (D-aspartate) O-methyltransferase involved in the repair of age-damaged proteins. J Hum Genet 44: 275–288, 1999 [DOI] [PubMed] [Google Scholar]
- 26.DeVry CG, Tsai W, and Clarke S. Structure of the human gene encoding the protein repair L-isoaspartyl (D-aspartyl) O-methyltransferase. Arch Biochem Biophys 335: 321–332, 1996 [DOI] [PubMed] [Google Scholar]
- 27.Doyle HA, Aswad DW, and Mamula MJ. Autoimmunity to isomerized histone H2B in systemic lupus erythematosus. Autoimmunity 46: 6–13, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle HA, Gee RJ, and Mamula MJ. A failure to repair self-proteins leads to T cell hyperproliferation and autoantibody production. J Immunol 171: 2840–2847, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Doyle HA, Gee RJ, and Mamula MJ. Altered immunogenicity of isoaspartate containing proteins. Autoimmunity 40: 131–137, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Doyle HA. and Mamula MJ. Posttranslational modifications of self-antigens. Ann N Y Acad Sci 1050: 1–9, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Doyle HA. and Mamula MJ. Autoantigenesis: the evolution of protein modifications in autoimmune disease. Curr Opin Immunol 24: 112–118, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, and Nystrom T. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci U S A 97: 5746–5749, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dzhambazov B, Holmdahl M, Yamada H, Lu S, Vestberg M, Holm B, Johnell O, Kihlberg J, and Holmdahl R. The major T cell epitope on type II collagen is glycosylated in normal cartilage but modified by arthritis in both rats and humans. Eur J Immunol 35: 357–366, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Eckert D, Biermann K, Nettersheim D, Gillis AJ, Steger K, Jack HM, Muller AM, Looijenga LH, and Schorle H. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev Biol 8: 106, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fedorova M, Bollineni RC, and Hoffmann R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev 33: 79–97, 2014 [DOI] [PubMed] [Google Scholar]
- 36.Frohnert BI. and Bernlohr DA. Protein carbonylation, mitochondrial dysfunction, and insulin resistance. Adv Nutr 4: 157–163, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gahring L, Carlson NG, Meyer EL, and Rogers SW. Granzyme B proteolysis of a neuronal glutamate receptor generates an autoantigen and is modulated by glycosylation. J Immunol 166: 1433–1438, 2001 [DOI] [PubMed] [Google Scholar]
- 38.Gergely P, Jr., Grossman C, Niland B, Puskas F, Neupane H, Allam F, Banki K, Phillips PE, and Perl A. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum 46: 175–190, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gergely P, Jr., Niland B, Gonchoroff N, Pullmann R, Jr., Phillips PE, and Perl A. Persistent mitochondrial hyperpolarization, increased reactive oxygen intermediate production, and cytoplasmic alkalinization characterize altered IL-10 signaling in patients with systemic lupus erythematosus. J Immunol 169: 1092–1101, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.German DC, Bloch CA, and Kredich NM. Measurements of S-adenosylmethionine and L-homocysteine metabolism in cultured human lymphoid cells. J Biol Chem 258: 10997–11003, 1983 [PubMed] [Google Scholar]
- 41.Goudarzi M, Chauthe S, Strawn SJ, Weber WM, Brenner DJ, and Fornace AJ. Quantitative metabolomic analysis of urinary citrulline and calcitroic acid in mice after exposure to various types of ionizing radiation. Int J Mol Sci 17, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta CR, Ratna JV, and Mohammad Y. Protein carbonylation as biomarker(s) in serum patients with type 2 diabetes. J Pharm Res 4: 348–351, 2011 [Google Scholar]
- 43.Harvey BP, Gee RJ, Haberman AM, Shlomchik MJ, and Mamula MJ. Antigen presentation and transfer between B cells and macrophages. Eur J Immunol 37: 1739–1751, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Harvey BP, Quan TE, Rudenga BJ, Roman RM, Craft J, and Mamula MJ. Editing antigen presentation: antigen transfer between human B lymphocytes and macrophages mediated by class A scavenger receptors. J Immunol 181: 4043–4051, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayem G, Nicaise-Roland P, Palazzo E, de Bandt M, Tubach F, Weber M, and Meyer O. Anti-oxidized low-density-lipoprotein (OxLDL) antibodies in systemic lupus erythematosus with and without antiphospholipid syndrome. Lupus 10: 346–351, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Hepburn AL, Lampert IA, Boyle JJ, Horncastle D, Ng WF, Layton M, Vyse TJ, Botto M, and Mason JC. In vivo evidence for apoptosis in the bone marrow in systemic lupus erythematosus. Ann Rheum Dis 66: 1106–1109, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann M, Voll RE, Zoller OM, Hagenhofer M, Ponner BB, and Kalden JR. Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum 41: 1241–1250, 1998 [DOI] [PubMed] [Google Scholar]
- 48.Heyn H. and Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13: 679–692, 2012 [DOI] [PubMed] [Google Scholar]
- 49.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, and Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol 171: 538–541, 2003 [DOI] [PubMed] [Google Scholar]
- 50.Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, Zhang G, Zhou Y, Su Y, and Lu Q. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol 35: 804–810, 2008 [PubMed] [Google Scholar]
- 51.Huang S, Litt M, and Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev 19: 1885–1893, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huebscher KJ, Lee J, Rovelli G, Ludin B, Matus A, Stauffer D, and Furst P. Protein isoaspartyl methyltransferase protects from Bax-induced apoptosis. Gene 240: 333–341, 1999 [DOI] [PubMed] [Google Scholar]
- 53.Infantino S, Benz B, Waldmann T, Jung M, Schneider R, and Reth M. Arginine methylation of the B cell antigen receptor promotes differentiation. J Exp Med 207: 711–719, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ireland JM. and Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med 208: 2625–2632, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwasaki H, Kovacic JC, Olive M, Beers JK, Yoshimoto T, Crook MF, Tonelli LH, and Nabel EG. Disruption of protein arginine N-methyltransferase 2 regulates leptin signaling and produces leanness in vivo through loss of STAT3 methylation. Circ Res 107: 992–1001, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson BA. and Aswad DW. Fragmentation of isoaspartyl peptides and proteins by carboxypeptidase Y: release of isoaspartyl dipeptides as a result of internal and external cleavage. Biochemistry 29: 4373–4380, 1990 [DOI] [PubMed] [Google Scholar]
- 57.Juarez M, Bang H, Hammar F, Reimer U, Dyke B, Sahbudin I, Buckley CD, Fisher B, Filer A, and Raza K. Identification of novel antiacetylated vimentin antibodies in patients with early inflammatory arthritis. Ann Rheum Dis 75: 1099–1107, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, Friday S, Li S, Patel RM, Subramanian V, Thompson P, Chen P, Fox DA, Pennathur S, and Kaplan MJ. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med 5: 178ra40, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kidd BA, Ho PP, Sharpe O, Zhao X, Tomooka BH, Kanter JL, Steinman L, and Robinson WH. Epitope spreading to citrullinated antigens in mouse models of autoimmune arthritis and demyelination. Arthritis Res Ther 10: R119, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, Richie E, and Bedford MT. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem 279: 25339–25344, 2004 [DOI] [PubMed] [Google Scholar]
- 61.Kinloch A, Tatzer V, Wait R, Peston D, Lundberg K, Donatien P, Moyes D, Taylor PC, and Venables PJ. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res Ther 7: R1421–R1429, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klareskog L, Ronnelid J, Lundberg K, Padyukov L, and Alfredsson L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol 26: 651–675, 2008 [DOI] [PubMed] [Google Scholar]
- 63.Kleinschmidt MA, Streubel G, Samans B, Krause M, and Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res 36: 3202–3213, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohse K, Wang Q, Stritzke S, Konigshoff M, Eickelberg O, and Yildirim AO. Protein arginine methyltransferases (prmt) are involved in Th17 cell differentiation. Am J Respir Crit Care Med 183: Abstract 4399, 2011 [Google Scholar]
- 65.Kuhn A, Herrmann M, Kleber S, Beckmann-Welle M, Fehsel K, Martin-Villalba A, Lehmann P, Ruzicka T, Krammer PH, and Kolb-Bachofen V. Accumulation of apoptotic cells in the epidermis of patients with cutaneous lupus erythematosus after ultraviolet irradiation. Arthritis Rheum 54: 939–950, 2006 [DOI] [PubMed] [Google Scholar]
- 66.Lammi A, Arikoski P, Simell S, Kinnunen T, Simell V, Paavanen-Huhtala S, Hinkkanen A, Veijola R, Knip M, Toppari J, Vaarala O, Simell O, and Ilonen J. Antibodies to deamidated gliadin peptide in diagnosis of celiac disease in children. J Pediatr Gastroenterol Nutr 60: 626–631, 2015 [DOI] [PubMed] [Google Scholar]
- 67.Lartigue A, Drouot L, Jouen F, Charlionet R, Tron F, and Gilbert D. Association between anti-nucleophosmin and anti-cardiolipin antibodies in (NZW x BXSB)F1 mice and human systemic lupus erythematosus. Arthritis Res Ther 7: R1394–R1403, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, and Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell 31: 212–221, 2008 [DOI] [PubMed] [Google Scholar]
- 69.Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, and Pestka S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem 280: 3656–3664, 2005 [DOI] [PubMed] [Google Scholar]
- 70.Lehmann PV, Forsthuber T, Miller A, and Sercarz EE. Spreading of T-cell autoimmunity to cryptic determinants of an autoantigen. Nature 358: 155–157, 1992 [DOI] [PubMed] [Google Scholar]
- 71.Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, and Huang S. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 115: 2028–2037, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K, Shen Q, Liu X, Zhu X, Li N, Cheng Z, Fan G, Wang Q, and Cao X. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat Immunol 17: 806–815, 2016 [DOI] [PubMed] [Google Scholar]
- 73.Lin WJ, Gary JD, Yang MC, Clarke S, and Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem 271: 15034–15044, 1996 [DOI] [PubMed] [Google Scholar]
- 74.Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, and Robertson KD. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res 32: 598–610, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu CL, Tangsombatvisit S, Rosenberg JM, Mandelbaum G, Gillespie EC, Gozani OP, Alizadeh AA, and Utz PJ. Specific post-translational histone modifications of neutrophil extracellular traps as immunogens and potential targets of lupus autoantibodies. Arthritis Res Ther 14: R25, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lowenson JD, Kim E, Young SG, and Clarke S. Limited accumulation of damaged proteins in l-isoaspartyl (D-aspartyl) O-methyltransferase-deficient mice. J Biol Chem 276: 20695–20702, 2001 [DOI] [PubMed] [Google Scholar]
- 77.Lu Q. The critical importance of epigenetics in autoimmunity. J Autoimmun 41: 1–5, 2013 [DOI] [PubMed] [Google Scholar]
- 78.Machold KP, Stamm TA, Nell VP, Pflugbeil S, Aletaha D, Steiner G, Uffmann M, and Smolen JS. Very recent onset rheumatoid arthritis: clinical and serological patient characteristics associated with radiographic progression over the first years of disease. Rheumatology (Oxford) 46: 342–349, 2007 [DOI] [PubMed] [Google Scholar]
- 79.Mamula MJ. The inability to process a self peptide allows T cells to escape tolerance. J Exp Med 177: 567–571, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mamula MJ. Lupus autoimmunity: from peptides to particles. Immunol Rev 144: 301–314, 1995 [DOI] [PubMed] [Google Scholar]
- 81.Mamula MJ. Epitope spreading: the role of self peptides and autoantigen processing by B lymphocytes. Immunol Rev 164: 231–239, 1998 [DOI] [PubMed] [Google Scholar]
- 82.Mamula MJ, Gee RJ, Elliot JI, Sette A, Southwood S, Jones P, and Blier PR. Isoaspartyl post-translational modification triggers autoimmune responses to self-proteins. J Biol Chem 274: 22321–22327, 1999 [DOI] [PubMed] [Google Scholar]
- 83.Mannering SI, Harrison LC, Williamson NA, Morris JS, Thearle DJ, Jensen KP, Kay TW, Rossjohn J, Falk BA, Nepom GT, and Purcell AW. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med 202: 1191–1197, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manoury B, Mazzeo D, Fugger L, Viner N, Ponsford M, Streeter H, Mazza G, Wraith DC, and Watts C. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat Immunol 3: 169–174, 2002 [DOI] [PubMed] [Google Scholar]
- 85.Marini JC. Interrelationships between glutamine and citrulline metabolism. Curr Opin Clin Nutr Metab Care 19: 62–66, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marre ML, James EA, and Piganelli JD. beta cell ER stress and the implications for immunogenicity in type 1 diabetes. Front Cell Dev Biol 3: 67, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marre ML, Profozich JL, Coneybeer JT, Geng X, Bertera S, Ford MJ, Trucco M, and Piganelli JD. Inherent ER stress in pancreatic islet beta cells causes self-recognition by autoreactive T cells in type 1 diabetes. J Autoimmun 72: 33–46, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, and Serre G. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J Immunol 166: 4177–4184, 2001 [DOI] [PubMed] [Google Scholar]
- 89.Matsuo K, Xiang Y, Nakamura H, Masuko K, Yudoh K, Noyori K, Nishioka K, Saito T, and Kato T. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res Ther 8: R175, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA, Hoffmuller U, Baron U, Olek S, Bluestone JA, and Brusko TM. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol 186: 3918–3926, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGinty JW, Chow IT, Greenbaum C, Odegard J, Kwok WW, and James EA. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes 63: 3033–3040, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McGinty JW, Marre ML, Bajzik V, Piganelli JD, and James EA. T cell epitopes and post-translationally modified epitopes in type 1 diabetes. Curr Diab Rep 15: 90, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McLaughlin RJ, de Haan A, Zaldumbide A, de Koning EJ, de Ru AH, van Veelen PA, van Lummel M, and Roep BO. Human islets and dendritic cells generate post-translationally modified islet autoantigens. Clin Exp Immunol 185: 133–140, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meissner T, Krause E, Lodige I, and Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell 119: 587–589; discussion 589–590, 2004 [DOI] [PubMed] [Google Scholar]
- 95.Meng X, Ezzati P, Smolik I, Bernstein CN, Hitchon CA, and El-Gabalawy HS. Characterization of autoantigens targeted by anti-citrullinated protein antibodies in vivo: prominent role for epitopes derived from histone 4 proteins. PLoS One 11: e0165501, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, and Guccione E. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol 19: 136–144, 2012 [DOI] [PubMed] [Google Scholar]
- 97.Miranda TB, Miranda M, Frankel A, and Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem 279: 22902–22907, 2004 [DOI] [PubMed] [Google Scholar]
- 98.Monneaux F, Lozano JM, Patarroyo ME, Briand JP, and Muller S. T cell recognition and therapeutic effect of a phosphorylated synthetic peptide of the 70K snRNP protein administered in MR/lpr mice. Eur J Immunol 33: 287–296, 2003 [DOI] [PubMed] [Google Scholar]
- 99.Morales Y, Nitzel DV, Price OM, Gui S, Li J, Qu J, and Hevel JM. Redox control of protein arginine methyltransferase 1 (PRMT1) activity. J Biol Chem 290: 14915–14926, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moss CX, Matthews SP, Lamont DJ, and Watts C. Asparagine deamidation perturbs antigen presentation on class II major histocompatibility complex molecules. J Biol Chem 280: 18498–18503, 2005 [DOI] [PubMed] [Google Scholar]
- 101.Moulton VR. and Tsokos GC. Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res Ther 13: 207, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mowen KA, Schurter BT, Fathman JW, David M, and Glimcher LH. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol Cell 15: 559–571, 2004 [DOI] [PubMed] [Google Scholar]
- 103.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, and David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 104: 731–741, 2001 [DOI] [PubMed] [Google Scholar]
- 104.Neugebauer KM, Merrill JT, Wener MH, Lahita RG, and Roth MB. SR proteins are autoantigens in patients with systemic lupus erythematosus. Importance of phosphoepitopes. Arthritis Rheum 43: 1768–1778, 2000 [DOI] [PubMed] [Google Scholar]
- 105.Nguyen H. and James EA. Immune recognition of citrullinated epitopes. Immunology 149: 131–138, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J 24: 1311–1317, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ospelt C, Bang H, Feist E, Camici GG, Keller S, Detert J, Kramer A, Gay S, Ghannam K, and Burmester GR. Carbamylation of vimentin is inducible by smoking and represents an independent autoantigen in rheumatoid arthritis. Ann Rheum Dis 76: 1176–1183, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, and Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J 26: 3558–3569, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parry RV. and Ward SG. Protein arginine methylation: a new handle on T lymphocytes? Trends Immunol 31: 164–169, 2010 [DOI] [PubMed] [Google Scholar]
- 110.Pritzker LB, Joshi S, Gowan JJ, Harauz G, and Moscarello MA. Deimination of myelin basic protein. 1. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry 39: 5374–5381, 2000 [DOI] [PubMed] [Google Scholar]
- 111.Qin J, Fu S, Speake C, Greenbaum CJ, and Odegard JM. NETosis-associated serum biomarkers are reduced in type 1 diabetes in association with neutrophil count. Clin Exp Immunol 184: 318–322, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rajpal G, Schuiki I, Liu M, Volchuk A, and Arvan P. Action of protein disulfide isomerase on proinsulin exit from endoplasmic reticulum of pancreatic beta-cells. J Biol Chem 287: 43–47, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raycroft MT, Harvey BP, Bruck MJ, and Mamula MJ. Inhibition of antigen trafficking through scavenger receptor A. J Biol Chem 287: 5310–5316, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Richard S, Morel M, and Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem J 388: 379–386, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodriguez-Calvo T, Zapardiel-Gonzalo J, Amirian N, Castillo E, Lajevardi Y, Krogvold L, Dahl-Jorgensen K, and von Herrath MG. Increase in pancreatic proinsulin and preservation of beta-cell mass in autoantibody-positive donors prior to type 1 diabetes onset. Diabetes 66: 1334–1345, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rondas D, Crevecoeur I, D'Hertog W, Ferreira GB, Staes A, Garg AD, Eizirik DL, Agostinis P, Gevaert K, Overbergh L, and Mathieu C. Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes 64: 573–586, 2015 [DOI] [PubMed] [Google Scholar]
- 117.Rui J, Deng S, Arazi A, Perdigoto AL, Liu Z, and Herold KC. Beta cells that resist immunological attack develop during progression of autoimmune diabetes in NOD mice. Cell Metab 25: 727–738, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rui J, Deng S, Lebastchi J, Clark PL, Usmani-Brown S, and Herold KC. Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia 59: 1021–1029, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ruskovska T. and Bernlohr DA. Oxidative stress and protein carbonylation in adipose tissue—implications for insulin resistance and diabetes mellitus. J Proteomics 92: 323–334, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanchez-Margalet V. and Najib S. p68 Sam is a substrate of the insulin receptor and associates with the SH2 domains of p85 PI3K. FEBS Lett 455: 307–310, 1999 [DOI] [PubMed] [Google Scholar]
- 121.Shacter E. Quantification and significance of protein oxidation in biological samples. Drug Metab Rev 32: 307–326, 2000 [DOI] [PubMed] [Google Scholar]
- 122.Shao CH, Capek HL, Patel KP, Wang M, Tang K, DeSouza C, Nagai R, Mayhan W, Periasamy M, and Bidasee KR. Carbonylation contributes to SERCA2a activity loss and diastolic dysfunction in a rat model of type 1 diabetes. Diabetes 60: 947–959, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Strickland FM. and Richardson BC. Epigenetics in human autoimmunity. Epigenetics in autoimmunity—DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity 41: 278–286, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Styskal J, Van Remmen H, Richardson A, and Salmon AB. Oxidative stress and diabetes: what can we learn about insulin resistance from antioxidant mutant mouse models? Free Radic Biol Med 52: 46–58, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun Q, Yang X, Zhong B, Jiao F, Li C, Li D, Lan X, Sun J, and Lu S. Upregulated protein arginine methyltransferase 1 by IL-4 increases eotaxin-1 expression in airway epithelial cells and participates in antigen-induced pulmonary inflammation in rats. J Immunol 188: 3506–3512, 2012 [DOI] [PubMed] [Google Scholar]
- 126.Suzuki A, Yamada R, Ohtake-Yamanaka M, Okazaki Y, Sawada T, and Yamamoto K. Anti-citrullinated collagen type I antibody is a target of autoimmunity in rheumatoid arthritis. Biochem Biophys Res Commun 333: 418–426, 2005 [DOI] [PubMed] [Google Scholar]
- 127.Tas SW, Quartier P, Botto M, and Fossati-Jimack L. Macrophages from patients with SLE and rheumatoid arthritis have defective adhesion in vitro, while only SLE macrophages have impaired uptake of apoptotic cells. Ann Rheum Dis 65: 216–221, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tucci M, Stucci S, Strippoli S, and Silvestris F. Cytokine overproduction, T-cell activation, and defective T-regulatory functions promote nephritis in systemic lupus erythematosus. J Biomed Biotechnol 2010: 457146, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Utz PJ. and Anderson P. Life and death decisions: regulation of apoptosis by proteolysis of signaling molecules. Cell Death Differ 7: 589–602, 2000 [DOI] [PubMed] [Google Scholar]
- 130.van Lummel M, Duinkerken G, van Veelen PA, de Ru A, Cordfunke R, Zaldumbide A, Gomez-Tourino I, Arif S, Peakman M, Drijfhout JW, and Roep BO. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 63: 237–247, 2014 [DOI] [PubMed] [Google Scholar]
- 131.van Stipdonk MJ, Willems AA, Amor S, Persoon-Deen C, Travers PJ, Boog CJ, and van Noort JM. T cells discriminate between differentially phosphorylated forms of alphaB-crystallin, a major central nervous system myelin antigen. Int Immunol 10: 943–950, 1998 [DOI] [PubMed] [Google Scholar]
- 132.Vehik K, Beam CA, Mahon JL, Schatz DA, Haller MJ, Sosenko JM, Skyler JS, and Krischer JP. Development of autoantibodies in the TrialNet Natural History Study. Diabetes Care 34: 1897–1901, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, and Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24: 179–189, 2006 [DOI] [PubMed] [Google Scholar]
- 134.Verheul MK, Yee A, Seaman A, Janssen GM, van Veelen PA, Drijfhout JW, Toes RE, Mahler M, and Trouw LA. Identification of carbamylated alpha 1 anti-trypsin (A1AT) as an antigenic target of anti-CarP antibodies in patients with rheumatoid arthritis. J Autoimmun 80: 77–84, 2017 [DOI] [PubMed] [Google Scholar]
- 135.Wagner AM, Cloos P, Bergholdt R, Boissy P, Andersen TL, Henriksen DB, Christiansen C, Christgau S, Pociot F, and Nerup J. Post-translational protein modifications in type 1 diabetes: a role for the repair enzyme protein-L-isoaspartate (D-aspartate) O-methyltransferase? Diabetologia 50: 676–681, 2007 [DOI] [PubMed] [Google Scholar]
- 136.Wagner AM, Cloos P, Bergholdt R, Eising S, Brorsson C, Stalhut M, Christgau S, Nerup J, and Pociot F. Posttranslational protein modifications in type 1 diabetes—genetic studies with PCMT1, the repair enzyme protein isoaspartate methyltransferase (PIMT) encoding gene. Rev Diabet Stud 5: 225–231, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, and Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293: 853–857, 2001 [DOI] [PubMed] [Google Scholar]
- 138.Wang L, Pal S, and Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol 28: 6262–6277, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y, Xiao Y, Zhong L, Ye D, Zhang J, Tu Y, Bornstein SR, Zhou Z, Lam KS, and Xu A. Increased neutrophil elastase and proteinase 3 and augmented NETosis are closely associated with beta-cell autoimmunity in patients with type 1 diabetes. Diabetes 63: 4239–4248, 2014 [DOI] [PubMed] [Google Scholar]
- 140.Weber S, Maass F, Schuemann M, Krause E, Suske G, and Bauer UM. PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes Dev 23: 118–132, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 141.Wegner N, Lundberg K, Kinloch A, Fisher B, Malmstrom V, Feldmann M, and Venables PJ. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunol Rev 233: 34–54, 2010 [DOI] [PubMed] [Google Scholar]
- 142.Wiik AS, van Venrooij WJ, and Pruijn GJ. All you wanted to know about anti-CCP but were afraid to ask. Autoimmun Rev 10: 90–93, 2010 [DOI] [PubMed] [Google Scholar]
- 143.Wolos JA, Frondorf KA, Davis GF, Jarvi ET, McCarthy JR, Bowlin TL. Selective inhibition of T cell activation by an inhibitor of S-adenosyl-L-homocysteine hydrolase. J Immunol 150: 3264–3273, 1993 [PubMed] [Google Scholar]
- 144.Wysocka J, Allis CD, and Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci 11: 344–355, 2006 [DOI] [PubMed] [Google Scholar]
- 145.Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, Zheng Y, Nishinakamura R, Xu GL, Wang H, Chen S, Gao S, and Zhu B. Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol Cell 64: 913–925, 2016 [DOI] [PubMed] [Google Scholar]
- 146.Yang L, Tan D, and Piao H. Myelin basic protein citrullination in multiple sclerosis: a potential therapeutic target for the pathology. Neurochem Res 41: 1845–1856, 2016 [DOI] [PubMed] [Google Scholar]
- 147.Yang ML, Doyle HA, Gee RJ, Lowenson JD, Clarke S, Lawson BR, Aswad DW, and Mamula MJ. Intracellular protein modification associated with altered T cell functions in autoimmunity. J Immunol 177: 4541–4549, 2006 [DOI] [PubMed] [Google Scholar]
- 148.Young GW, Hoofring SA, Mamula MJ, Doyle HA, Bunick GJ, Hu Y, and Aswad DW. Protein L-isoaspartyl methyltransferase catalyzes in vivo racemization of Aspartate-25 in mammalian histone H2B. J Biol Chem 280: 26094–26098, 2005 [DOI] [PubMed] [Google Scholar]
- 149.Yu HC, Lai PH, Lai NS, Huang HB, Koo M, and Lu MC. Increased serum levels of anti-carbamylated 78-kDa glucose-regulated protein antibody in patients with rheumatoid arthritis. Int J Mol Sci 17:1510–1518, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yuan S, Patel RP, and Kevil CG. Working with nitric oxide and hydrogen sulfide in biological systems. Am J Physiol Lung Cell Mol Physiol 308: L403–L415, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zamvil SS, Mitchell DJ, Moore AC, Kitamura K, Steinman L, and Rothbard JB. T-cell epitope of the autoantigen myelin basic protein that induces encephalomyelitis. Nature 324: 258–260, 1986 [DOI] [PubMed] [Google Scholar]
- 152.Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, Luo J, Yen RC, Li Y, and Baylin SB. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol Cell 65: 323–335, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zullo A, Sommese L, Nicoletti G, Donatelli F, Mancini FP, and Napoli C. Epigenetics and type 1 diabetes: mechanisms and translational applications. Transl Res 185: 85–93, 2017 [DOI] [PubMed] [Google Scholar]