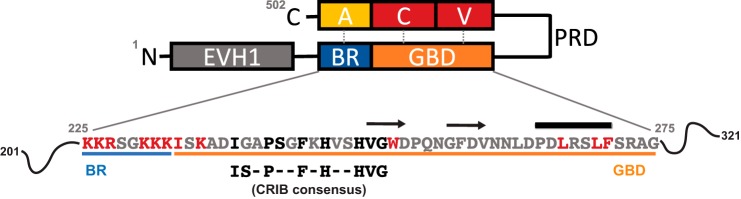
Figure 1.
Domain architecture of WASP. Structural domains and regions with assigned function of WASP are highlighted as follows: the Ena/VASP homology domain 1 (EVH1, gray); the basic region (BR, blue); the G protein-binding domain (GBD, orange); the central proline-rich domain (PRD); the verprolin homology (V, red); central (C, red); and acidic region (A, yellow). WASP exists in an autoinhibited conformation when not bound by effectors, and the BR and A regions and the CV and GBD regions form contacts that prevent the VCA region binding and activating Arp2/3. Only WASP residues 225–275, which include the BR and residues of the GBD mutated in this study, are expanded to show their sequence below, and the residues selected for mutagenesis are highlighted. Residues colored black are CRIB consensus residues, which were all subject to mutation in this study; residues outside the CRIB region mutated in this work are colored red. The CRIB consensus sequence is also included. Secondary structure elements in WASP when bound to Cdc42 are shown above the sequence. β-Strands are denoted by arrows and the helix by a cylinder. The limits of the secondary structure elements are taken from Ref. 14 with amendments included from Ref. 29. The accession number for WASP is UniProt no. P42768.