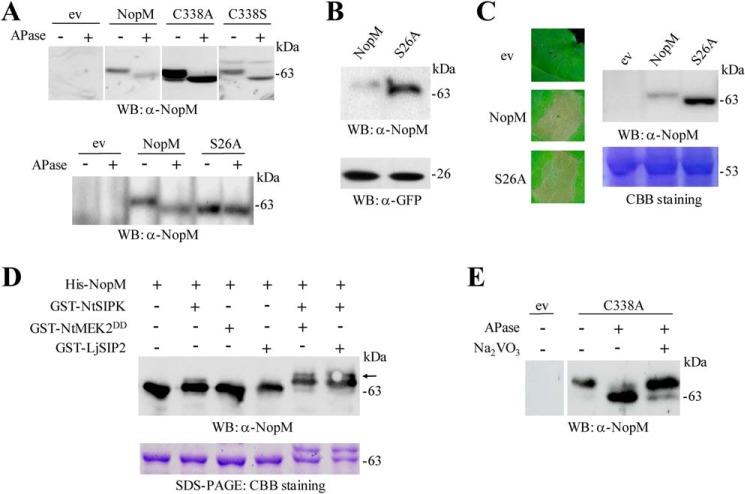
Figure 6.
Phosphorylation of NopM in planta and in vitro. A, analysis of NopM and indicated variants isolated from tobacco leaves transformed with A. tumefaciens. Agrobacteria carrying the empty vector (ev) were used as negative control. Soluble proteins were extracted from the transformed leaf tissue, and aliquots were treated with APase. Samples were then subjected to Western blot (WB) analysis with anti-NopM antibodies. B, Western blot analysis of NopM and the S26A variant expressed in tobacco leaves. For comparison, co-expressed GFP was also analyzed in the obtained protein extracts. C, expression of the S26A variant in tobacco induces cell death. NopM expression and empty vector controls were used for comparison. The pictures were taken 3 days after infiltration with agrobacteria. A corresponding Western blot confirmed expression of the proteins. Coomassie Brilliant Blue (CBB) staining of ribulose-bisphosphate carboxylase/oxygenase large subunit in a parallel gel indicated the use of equal amounts of proteins. D, in vitro phosphorylation of NopM by NtSIPK. Indicated proteins with GST or His tags were expressed in E. coli and purified by affinity chromatography. The MAPK kinases NtMEK2DD and LjSIP2 were used for activation of NtSIPK. Proteins were incubated in phosphorylation buffer containing ATP for 30 min at 30 °C. The samples were analyzed by Western blotting using an anti-NopM antibody. NopM phosphorylated by activated NtSIPK migrated more slowly on the gel (upper bands marked by an arrow). A parallel protein gel was stained with Coomassie Brilliant Blue. E, analysis of the C338A variant expressed in L. japonicus roots. Soluble proteins were extracted 28 days after transformation, and aliquots were treated with APase and 10 mm Na3VO4. Samples were then subjected to Western blot analysis with anti-NopM antibodies.