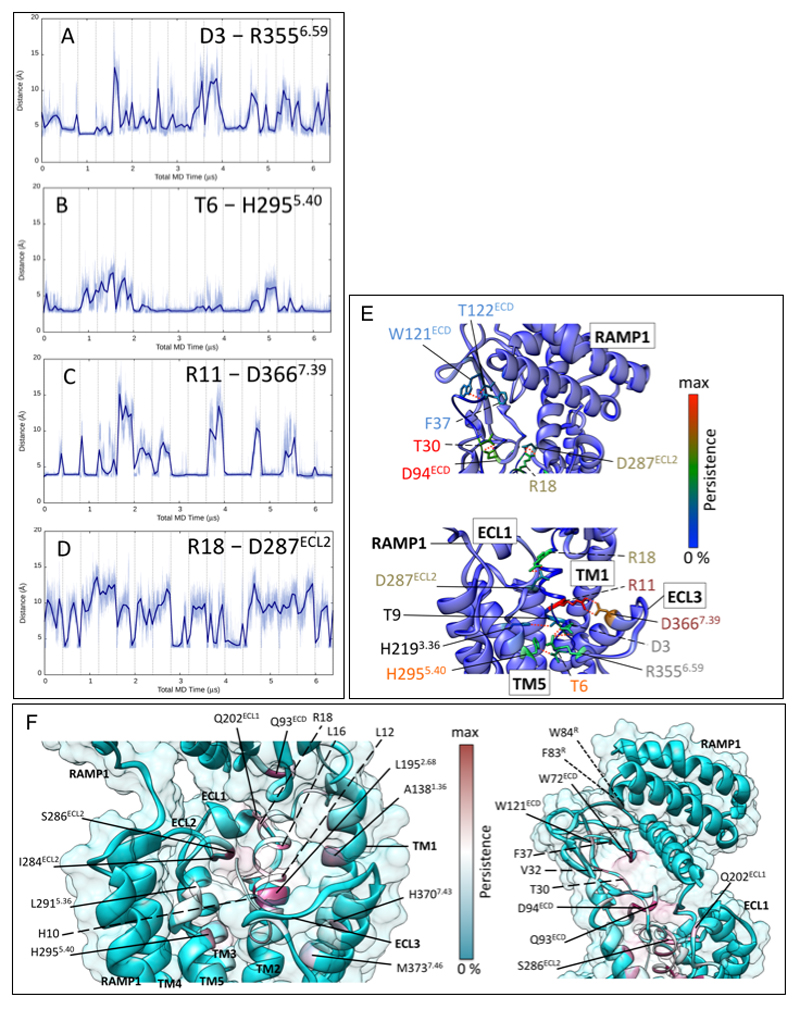
Extended Data Figure 9. CGRP makes extensive stable interactions with CLR.
A-D; Distances between CGRP and CLR residues relevant to key hydrogen bonds. The x-axis denotes sampling time for the 16 merged MD replicas of the whole system (each replica is separated by vertical dashed lines). A; Distance between the peptide D3P carboxylic carbon and receptor R3556.59 guanidinium carbon. B; distance between the peptide T6P side chain oxygen atom and the receptor H2955.40 side chain nitrogen atoms (for each frame, the closest nitrogen to T6P was considered). C; Distance between the peptide R11P guanidinium carbon and the receptor D3667.39 carboxylic carbon. D; Distance between peptide R18P guanidinium carbon and receptor D287ECL2 carboxylic carbon. In most cases the distances corresponding to hydrogen bond formation are slightly longer than the standard 2.8 Å. E; H-bonds between CGRP and CLR during MD simulations (6.4 μs). The total persistency of a residue side chain is plotted onto the experimental structure according to a rainbow colour scale, with residues never involved in blue and residues highly involved in red. The peptide (italics, dashed line) is depicted as thin ribbon, while the receptor (solid line) is shown as bulky ribbon. Key side chains are shown, but for intermittent H-bonds the rotameric state has been modified to show an interaction. Residues forming an interaction network are labelled with the same colour. Lower panel, H-bonds between the CGRP N-terminus and the TM bundle of CLR. Upper panel, H-bonds between the CGRP C-terminus and the ECD of CLR; quantitative data on the persistence of H-bonds during the simulations are reported in Supp. Information Table 3. F; Contacts between CGRP and CLR / RAMP1 during MD simulations (6.4 μs). The total persistency of a residue side chain is plotted onto the experimental structure according to a cyan-maroon colour scale, with residues never involved in cyan and residue highly involved in maroon. The peptide (italics, dashed line) is depicted as a thin ribbon, while the receptor (solid line) is shown as a bulky ribbon and transparent surface. Left, contacts between the N-terminus of CGRP and the TM bundle of the CLR: highly persistent hydrophobic interactions characterize peptide residues L12P, L16P, H10P and receptor residues L1952.68, A1381.36 and H2955.40. Right, contacts between the C-terminus of CGRP and the ECD of CLR; highly persistent contacts characterize peptide residues V32P, T30P, F37P and receptor residues Q93ECD and W72ECD. RAMP1 residues F83R, W84R are mainly engaged by CGRP residue F37P.