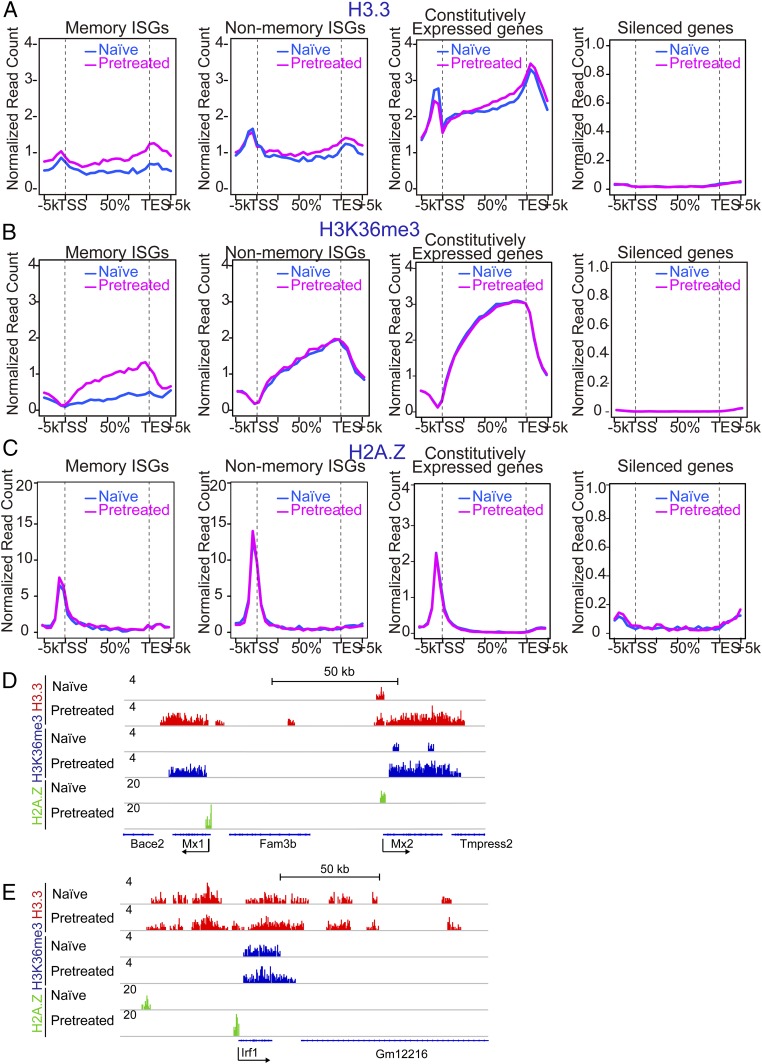
Fig. 4.
Memory ISGs are marked by the histones H3.3 and H3K36me3. (A–C) ChIP-seq analysis was performed for genome-wide distribution of H3.3 (A), H3K36me3 (B), and H2AZ (C) over memory ISGs, nonmemory ISGs (Left), and constitutively expressed or silent genes (Right) in naïve or pretreated MEFs without restimulation. Pretreated cells were left without IFN for 48 h. See SI Appendix, Fig. S4 D–H for IGV examples. (D) IGV images of H3.3, H3K36me3, and H2AZ peaks over memory ISGs (Mx1 and Mx2) in naïve and pretreated cells. (E) IGV images of H3.3, H3K36me3, and H2AZ peaks over nonmemory ISG (Irf1) in naïve and pretreated cells.