Abstract
Elucidation of both the molecular composition and organization of the ocular lens is a prerequisite to understand its development, function, pathology, regenerative capacity, as well as to model lens development and disease using in vitro differentiation of pluripotent stem cells. Lens is comprised of the anterior lens epithelium and posterior lens fibers, which form the bulk of the lens. Lens fibers differentiate from lens epithelial cells through cell cycle exit-coupled differentiation that includes cellular elongation, accumulation of crystallins, cytoskeleton and membrane remodeling, and degradation of organelles within the central region of the lens. Here, we profiled spatiotemporal expression dynamics of both mRNAs and non-coding RNAs from microdissected mouse nascent lens epithelium and lens fibers at four developmental time points (embryonic [E] day 14.5, E16.5, E18.5, and P0.5) by RNA-seq. During this critical time window, multiple complex biosynthetic and catabolic processes generate the molecular and structural foundation for lens transparency. Throughout this developmental window, 3,544 and 3,518 genes show consistently and significantly greater expression in the nascent lens epithelium and fibers, respectively. Comprehensive data analysis confirmed major roles of FGF-MAPK, Wnt/β-catenin, PI3K/AKT, TGF-β, and BMP signaling pathways and revealed significant novel contributions of mTOR, EIF2, EIF4 and p70S6K signaling in lens formation. Unbiased motif analysis within promoter regions of these genes with consistent expression changes between epithelium and fiber cells revealed an enrichment for both established (e.g. E2Fs, Etv5, Hsf4, c-Maf, MafG, MafK, N-Myc, and Pax6) transcription factors and a number of novel regulators of lens formation, such as Arntl2, Dmrta2, Stat5a, Stat5b, and Tulp3. In conclusion, the present RNA-seq data serves as a comprehensive reference resource for deciphering molecular principles of normal mammalian lens differentiation, mapping a full spectrum of signaling pathways and DNA-binding transcription factors operating in both lens compartments, and predicting novel pathways required to establish lens transparency.
Keywords: Differentiation, lens epithelium, lens fiber, RNA-seq, transcription factors, mTOR signaling
1. Introduction
Ocular lens development represents an advantageous system to probe mechanisms of cell differentiation, cell remodeling, signaling, and transcriptional regulation, as positions of individual cells within the lens provides exact information about their differentiation status (Cvekl and Zhang, 2017; Lovicu and McAvoy, 2005; Piatigorsky, 1981). Lens is of an ectodermal origin; lens development becomes visible as the thickening of the head surface ectoderm, the lens placode, at E9.5 of mouse embryonic development (Chow and Lang,2001; Cvekl and Ashery-Padan, 2014; Lovicu and McAvoy, 2005). Invagination of the lens placode leads to the formation of the lens pit in which the ectodermal sheet of cells is converted into a transitional 3D structure, the lens vesicle (mouse E11 stage), comprised of lens precursor cells. Lens vesicle is a polarized structure; its posterior cells exit the cell cycle and undergo terminal differentiation to become the primary lens fibers that elongate towards the anterior portion of the lens (E12.5-E14.5). In parallel, the anterior cells of the lens vesicle differentiate into the nascent single layered cuboidal lens epithelium. From E14.5, lens growth is driven by cell division within the lens epithelium compartment through the formation of secondary lens fibers that add new concentric outer cell layers while the primary lens fibers are pushed inward into the lens “nucleus” (Bassnett and Sikic, 2017). Between E16.5-E18.5, the most inner lens fiber cells degrade their subcellular organelles, including the mitochondria, endoplasmic reticulum, Golgi apparatus, and nuclei, to form the organelle free zone (OFZ) (Bassnett, 2009). Lens fiber cell elongation extends the original lens length by a factor of 1,000-fold (Audette et al., 2017; Bassnett and Costello, 2017), while their cytoplasmic compartments accumulate crystallin proteins (Cvekl and Zhang, 2017). The lens fibers form an elaborate cytoskeletal and membrane organization to support lens transparency (Beyer and Berthoud, 2014; Dahm et al., 2011; Mathias et al., 2010; Schey et al., 2017; Song et al., 2009; Wang et al., 2016).
Although a number of important genes are known in the lens and their roles have been established (Graw, 2003; Hejtmancik et al., 2015), very little is known about the global expression of genes in the lens epithelium and lens fiber cell compartments as the nascent lens epithelium matures and lens fiber cells undergo terminal differentiation, organelle degradation, and maturation (Bassnett, 2009; Lovicu and McAvoy, 2005; Martinez and de Iongh, 2010). Previous longitudinal studies of lens transcriptomes typically employed whole lens (Kakrana et al., 2018) and microarrays, which were not suitable to quantify differences in mRNAs of highly expressed genes, such as crystallins (He et al., 2016; He et al., 2010; Pontoriero et al., 2008). In contrast, RNA-seq platform provides a better quantification of gene expression (Wilhelm et al., 2008), and the technique of lens micro-dissection to obtain epithelium and fiber makes it possible to quantify and compare spatial gene expression (Hoang et al., 2014; Sun et al., 2015a). RNA expression profiling studies in other differentiation models have revealed that spatiotemporally co-regulated genes are often regulated by overlapping sets of DNA-binding transcription factors and common signaling pathways (Bolouri, 2014; Soon et al., 2013). Computational tools have also been developed for unbiased analysis of c/s-regulatory elements in the promoters and enhancers of coregulated genes to predict cognate DNA-binding transcription factors (Bailey et al., 2009; Heinz et al., 2010).
Lens differentiation is regulated coordinately by a number of DNA-binding transcription factors (e.g. c-Maf, FoxE3, Gata3, Hsf4, Pax6, Prox1, and Sox1) and multiple canonical extracellular signaling pathways (Cvekl and Zhang, 2017; Lovicu and McAvoy, 2005). Amongst them, BMP and FGF signaling have been shown as the primary forces to regulate the cell cycle exit-coupled lens fiber cell differentiation program (Cvekl and Zhang, 2017; Griep, 2006). A combined action of BMP and FGF signaling drives both primary (Jarrin et al., 2012) and secondary lens fiber cell differentiation (Boswell and Musil, 2015; Boswell et al.2008). Additional novel components were recently incorporated into these pathways, including connexin 50 (Gja8) and E3 ubiquitin ligase Skp2 (Shi et al., 2015). It has been shown that FGF signaling regulates crystallin gene expression by three main TFs: c-Maf (Xie et al., 2016), Prox1(Audette et al., 2016), and Hsf4 (Hu and Mivechi, 2006). Both the Maf promoter and the Cryaa enhancer/promoter are regulated by direct binding of c-Jun and Etv5 (Xie et al., 2016), known targets of FGF signaling in other systems (Brewer et al., 2016; Guillemot and Zimmer, 2011; Turner and Grose, 2010). Expression of Mip (aquaporin 0) is also regulated by FGF signaling (Golestaneh et al., 2004). From these and other studies, a number of gene regulatory networks (GRNs) of lens cell formation (Anand and Lachke, 2017; Cvekl and Ashery-Padan, 2014) and crystallin gene expression (Cvekl and Zhang, 2017) have been constructed. Nevertheless, the full range and spectrum of lens differentiation- maturation processes and the underlying signaling pathways and TFs, especially at the level of nascent epithelium and fiber cell compartments, remain poorly understood.
Analysis of transcriptomes during lens differentiation, including gene loss-of-function studies, is a powerful method to reveal the dynamic molecular events of lens differentiation (Audette et al., 2016; Cavalheiro et al., 2017; He et al., 2010; Landgren et al., 2008; Shaham et al., 2013). From E14.5, it is possible to microdissect lenses into the epithelium and fiber cell mass and evaluate terminal differentiation of both nascent lens compartments. In the present study, we analyzed the expression of both mRNAs and non-coding RNAs (ncRNAs) at E14.5, E16.5, E18.5, and in newborn (P0.5) mouse lens epithelium and fiber compartments. We found novel waves of processes linked to differentiation in both lens compartments, dynamic changes in expression of established and new TFs. We also uncovered novel pathways and genes that control lens differentiation and tissue remodeling to support lens transparency. These data can be used for additional comparative studies with available chicken (Chauss et al., 2014) and human (Hawse et al., 2005; Hawse et al., 2004)and emerging lens transcriptome data from other model organisms, to enhance our understanding of lens development.
2. Materials and Methods
2.1. Tissue samples and RNA isolation
Mouse lenses from E14.5, E16.5, E18.5 and P0.5 CD1 mice (Charles River Laboratories) were removed from eyes and dissected under the microscope into lens epithelium and lens fiber as described earlier (Hoang et al., 2014; Zelenka et al., 2009). Epithelial and fiber fractions from multiple embryos were pooled into three biological replicates for each time point. 10 P0.5 lenses, 15 E18.5 lenses, 30 E16.5 lenses, and 50 E14.5 lenses were used for each biological replicate. Total RNA was extracted using Trizol. The purified RNAs were then treated with DnaseI (DNA-free™ DNA Removal Kit, Cat: AM1906) and RNA integrity was checked by bioanalyzer. The RNA samples with RNA integrity number (RIN) > 8 were used for RNA-seq library preparation and sequencing.
2.2. RNA-seq Library Preparation and Sequencing
We used KAPA RNA HyperPrep Kits with RiboErase (HMR) for RNA-seq library preparation according to the manufacturer’s instructions. The qualities and quantities of RNA-seq libraries were checked by bioanalyzer and qPCR. Subsequently, RNA-seq libraries were sent for 100bp paired-end sequencing on an Illumina HiSeq 2500 instrument in the Einstein Epigenomics facility.
2.3. Quantitative RT-PCR validation
Total RNA extracted as described above (2.1) was reverse transcribed into cDNA with ProtoScript II reverse Transcriptase (NEB: M0368) with Random Hexamer, following manufacturer’s instructions. The cDNA was diluted 5* and qPCR was conducted with Applied Biosystems (ABI Foster City, CA) 7900HT qPCR system and Power SYBR green (Invitrogen: 4367659) in 384-well plates. Since all known genes often used as reference for comparing different samples showed spatiotemporally differential expression based on our RNA-seq data (see below), we used splicing factor 3b subunit 1 Sf3b1, a gene expressed at a constant level, as the reference gene for normalization across samples. Student t test was performed to evaluate differential gene expression between epithelium and fiber at the same time point or between different time points of the same tissue. ANOVA was used to test differential gene expression between epithelium and fiber across all time points. Primers are listed in Supplementary Table S2.
2.4. RNA-seq Data Analysis
RNA-seq reads were aligned to the mouse genome (mm10) using Tophat (version 2.0.13) (Trapnell et al., 2009). The number of RNA-seq fragments mapped to each gene in the Refseq gene annotation (downloaded from the UCSC genome browser in 03/2017) was then counted using HTseq (version 0.6.1) (Anders et al., 2015). The Cuffdiff (version 2.2.1) (Trapnell et al., 2010) in the cufflinks package was also used to generate the normalized gene-expression level, FPKMs. Coding/UTR/intronic/intergenic rates and fragment size, were calculated by Picard for quality control (http://broadinstitute.github.io/picard). At the end,12,611 genes were considered lens expressed, based on their mean FPKM > 1 in any of the 8 sample groups (E14.5 epithelium, E14.5 fiber, E16.5 epithelium, E16.5 fiber, E18.5 epithelium, E18.5 fiber, P0.5 epithelium, P0.5 fiber), and selected for differential expression analysis and principle component analysis (PCA) by DESeq2 (Love et al., 2014). The software DAVID (version 6.8) (Huang da et al., 2009) was used for Gene Ontology (GO) analysis of the differentially expressed genes (DEGs), with the 12,611 expressed genes as background. Ingenuity pathway analysis (IPA version 01–10) was used for canonical pathway analysis, with the ingenuity knowledge base (genes only) as background. For GO term analysis and mouse phenotype analysis of temporal gene clusters in lens epithelium and lens fiber, Toppgenes (Chen et al., 2007) was also used. The RNA-seq data have been deposited in the Gene Expression Omnibus (GEO; GSE113887)
2.5. Time course gene expression and transition pattern analysis
To model the temporal changes in gene expression, we first collected all the differentially expressed genes between transition time points in epithelium and fiber. Then the transitional changes of each gene were categorized into up (+1), down (−1) and constant (0) based on the fold change (FC) at the adjacent time points (Ranked by E16.5 VS E14.5, E18.5 VS E16.5, P0.5 VS E18.5). Theoretically, there are 33=27 possible groups of transition patterns.
2.6. Unbiased analysis of TF expression
To predict novel and spatiotemporally enriched TFs in lens epithelium and fiber, 1484 mouse TFs were downloaded from the AnimalTFDB 2.0 (Zhang et al., 2015). The expression fold changes of these TFs in lens versus whole body (minus eye) comparisons were collected from the eye database iSyTE 2.0 (Kakrana et al., 2018). To predict their potential involvement in regulating eye development, we scanned the 2kb regions near the transcription start sites (TSS) of the DEGs with higher expression in either lens epithelium or lens fiber for enriched motifs using HOMER (v 4.7) (Heinz et al., 2010). The corresponding motif logos were downloaded from either the JASPAR database (Sandelin et al., 2004) or the Ismara database (Balwierz et al., 2014).
2.7. Predicting fractions of lens epithelium and fiber in whole lens gene expression data
Combining our RNA-seq data with the top lens enriched genes downloaded from the iSyTE 2.0, we selected 348 genes whose expression is lens enriched and is higher in either epithelium or fiber. We further excluded genes that are highly expressed in retina by comparing to retina RNA-seq data (GSE52006) to obtain a final list of 282 lens “signature genes”, including 65 lens epithelium signature genes and 217 lens fiber signature genes (Supplementary File S5, S6). We next generated a simulation expression value (S, in FPKM) for each of the signature genes as, S = α * (FPKM in epithelium) + (1-α) * (FPKM in fiber), where α is the relative percentage of epithelium in whole lens. We then calculated the Spearman’s correlation coefficient between the S and the observed expression value from whole lens for the 282 genes. The predicted percentage of epithelium is the α value that gives the highest correlation coefficient between simulation and observed data.
2.8. Co-expression analysis of ncRNAs and their adjacent genes
ncRNAs were identified by the “NR” in the IDs of the Refseq annotation, and those exhibiting temporal differential expression were analyzed for co-expression with flanking coding or noncoding genes (<100 kb separation on chromosomes by bedtools (v2.23.0)). Pearson’s correlation coefficients across four time points were calculated for each pair of non-coding gene and its adjacent gene(s). The resulted p values were adjusted for multiple test correction by the Benjamini-Hochberg method. At the end, we considered pairs with adjusted p value < 0.05 and correlation coefficient > 0.7 or < −0.7 as significantly co-expressed.
3. Results and Discussion
3.1. RNA-seq reveals profound differences between lens epithelium and fiber transcriptome
To analyze nascent lens epithelium and fiber cell differentiation, we microdissected E14.5, E16.5, E18.5 and P0.5 mouse lenses into epithelium and fiber compartments. We used RNA-seq to determine spatiotemporal gene expression profiles of lens development (Fig. 1A). We obtained on average 30 million RNA-seq read pairs per sample (Supplementary Table S1). A total of 12,611 genes were found expressed in embryonic lenses (mean FPKM > 1 in any of the eight sample groups). Differential expression analysis by DESeq2 identified a total of 5,805 genes with significant higher expression in lens epithelium and 5,617 higher in lens fiber (adjusted p-value (padj) <0.05 by Benjamini- Hochberg method) in at least one of the four stages. Among these spatially differentially expressed genes (sDEGs), 3,544 and 3,518 genes were expressed higher in epithelium and fiber consistently from E14.5 to P0.5, respectively (Fig. 1B). This analysis quantitatively demonstrates marked differences in nascent lens epithelium and fiber cell transcriptomes and provides a data source to explain phenotypic differences between these two cell types at the molecular level.
Fig. 1. Global view of spatial expression profiles of embryonic lens epithelium and fiber.
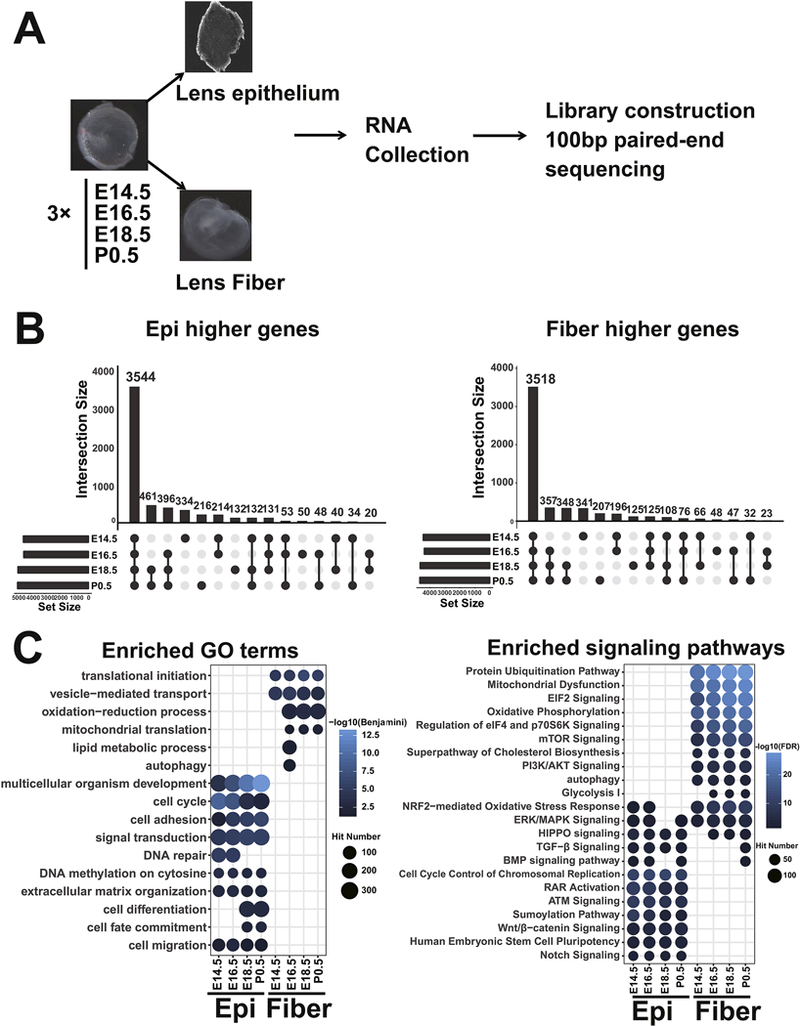
A. The workflow of RNA-seq data acquisition. B. Bar plots showing overlap of sDEGs at each time point. C. Enriched GO terms and signaling pathways of sDEGs. The GO terms and signaling pathways were first divided into fiber and epithelium (Epi) enriched and then ranked by their adjusted p values. The blue color corresponds to either -log10(Benjamini) or -log10(FDR). The dot size corresponds to the number of genes in the GO term or signaling pathways.
To gain initial insights into function of these sDEGs, we conducted GO term analysis and uncovered how specific signaling pathways are employed in lens epithelium and lens fibers. GO analysis with DAVID (Huang da et al., 2009) revealed “translational initiation” as the top enriched GO term in lens fiber up-regulated genes, followed by “vesicle-mediated transport”, ‘‘oxidation-reduction process”, “mitochondrial translation”, “lipid metabolic process” and “autophagy”. These findings are consistent with the known general processes implicated in fiber cell morphogenesis (Audette et al., 2017; McAvoy et al., 2017) (discussed below). The protein concentration in lens fibers reaches ~450 mg/ml (Bassnett and Costello, 2017); the mobilization of components of the translational apparatus in the lens fibers is evident from the present data. The translational initiation process requires the participation of multiple eukaryotic translation initiation complexes (eIF1-eIF6) (Kong and Lasko, 2012). Among the 54 eIF subunits expressed in lens, we noticed that 33 were expressed higher in fibers, including Eif3h, which was shown to regulate crystallin protein translation in zebrafish lens (Choudhuri et al., 2013). In contrast, only 3 Eifs were expressed higher in epithelium. The enrichment of Eifs in fiber was significant (Fisher exact test, p=1e-7), indicating higher translation activity in lens fiber cells. For genes expressed higher in nascent lens epithelium, five GO terms were notably enriched, including “multicellular organism development”, “cell cycle”, “cell adhesion”, “signal transduction”, and “DNA repair”, which are highly related to lens morphogenesis (Fig. 1C). It is also noteworthy that these terms are generally consistent with proliferating epithelia cell populations. Overall, this analysis shows that there are specific molecular processes uniquely employed by each of the two lens compartments.
Analysis of signaling pathways by IPA and their distribution also revealed pathways critical for the formation of lens epithelium and lens fibers (Fig. 1D). Many canonical signaling pathways in lens development are enriched in both epithelium and fiber higher expressed genes, including “ERK/MAPK Signaling” (Lovicu and McAvoy, 2001), “BMP signaling pathway”, “TGF-β signaling”, and “HIPPO signaling” (Cvekl and Zhang, 2017). In addition to these four pathways that are well established in lens development, the “NRF2-mediated oxidative stress response” term implicates redox-regulation in lens development, consistent with the formation of hypoxic conditions during lens embryogenesis (Baba et al., 2013).
Multiple signaling pathways were enriched only in genes of higher expression in the fiber cell compartment, such as the translation initiation associated signaling pathways including “Eif2 signaling”, “Regulation of eIF4 and p70S6K signaling”, and “mTOR Signaling,” consistent with the eIFs enrichment in lens fiber cells described above (Fig. 1D). Epithelium up-regulated genes were enriched in “Cell Cycle Control of Chromosomal Replication”, “RAR Activation”, ‘‘Notch Signaling”, and “Wnt/β-catenin Signaling”. These findings are consistent with both established (Bassnett and Sikic, 2017; Cvekl and Wang, 2009; Fujimura, 2016) and emerging (Mesa et al., 2016) themes in lens epithelial biology. Interestingly, we observed that genes expressed higher in lens fibers were enriched in “Protein Ubiquitination Pathway” but genes expressed higher in epithelium were enriched in “Sumoylation Pathway” (Fig. 1D), suggesting that different protein posttranslational modifications are preferentially used during early and more advanced stages of lens development.
To validate the RNA-seq findings, qRT-PCR analysis of the mRNAs from 12 genes encoding Eif3k, Mtor, Plekhml, Bnip3, Pax6, Tulp3, Stat5a, Sumol, Sumo3, Sae1, Uba2, and Kcnq1ot1, was conducted using independent cDNA preparations and Sf3b1 was used for data normalization. The results (Fig. 2) confirmed overall the observed difference in RNA-seq data, and these genes are discussed below.
Fig. 2. Quantitative RT-PCR validation of 12 differentially expressed mRNAs.
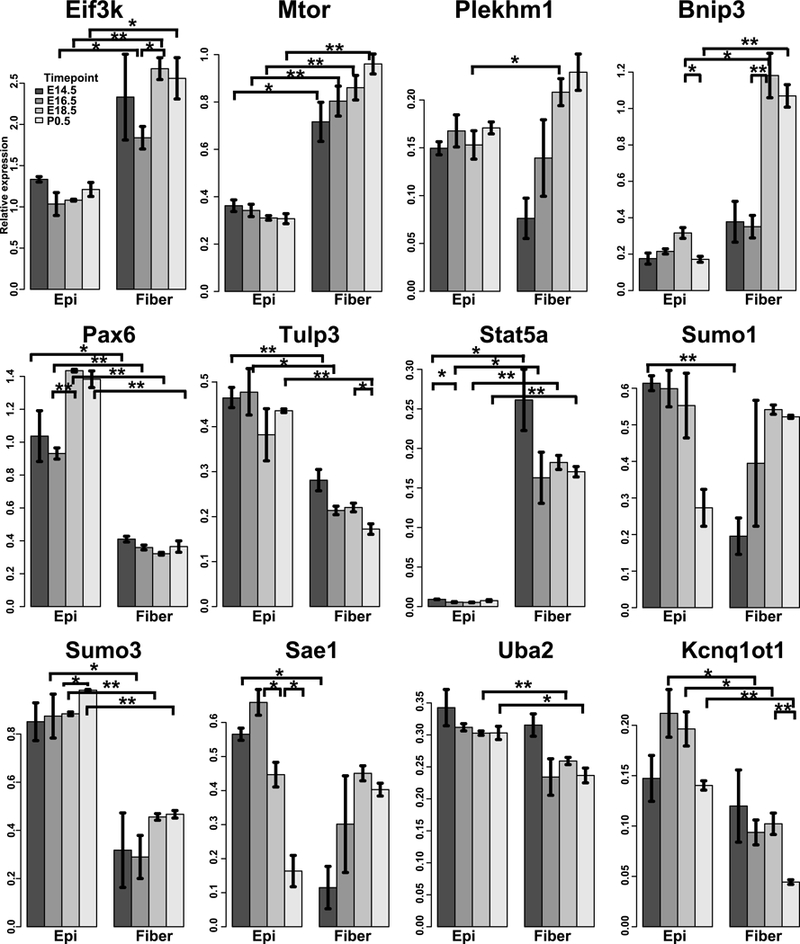
The barplots show the relative expression of the 12 genes normalized to the Sf3b1 in 8 samples. 3 biological replicates were used for each sample, except that 2 biological replicates were used for P1 Epi in some genes. “*” and “**” indicate p values (t-test) < 0.05 and < 0.01, respectively. Except for Plekhml and Sumol, all other genes were also significantly differentially expressed between epithelium and fiber across all time points by ANOVA tests.
3.2. Global temporal transcriptome dynamics in lens development
The temporally differentially expressed genes (tDEGs) between transition time points (E16.5 vs E14.5; E18.5 vs E16.5; P0.5 vs E18.5) were analyzed separately in epithelium and fiber using the DESeq2 (Fig. 2S A). A total of 4,324 and 2,923 genes (Fig. 11A) exhibited significant change (padj < 0.05) in epithelium and fiber, respectively. These tDEGs could be classified into 23 and 25 clusters in epithelium and fiber, respectively (Fig. 11A), which revealed for the first time several waves of gene expression during nascent lens epithelium and fiber differentiation (Supplementary table S3, S4).
As expected, compared to gene groups with expression changes in earlier stages (E16.5 vs E14.5, E18.5 vs E16.5), P0.5 vs E18.5 group had the fewest tDEGs (Fig. 2S).Using IPA, we found that signaling pathways “Oxidative Phosphorylation” and “Mitochondrial Dysfunction” were only enriched in genes with increased expression in lens fiber from E14.5 to P0.5, while “Sumoylation pathway” and “Notch Signaling” were only enriched in genes with significant temporal expression changes in epithelium. In addition, the canonical signaling pathways shared by epithelium and fiber were highlighted in the groups of genes with increased expression in both lens epithelium and fiber from E16.5 to E18.5, such as “FGF Signaling”, ‘BMP Signaling’, ‘TGF-beta Signaling’, and ‘RAR Signaling’. Interestingly, ‘P70S6K Signaling’, ‘EIF2 Signaling’, ‘PI3K/AKT Signaling’, ‘mTOR Signaling’, ‘Protein Ubiquitination Pathway’, ‘Cholesterol Biosynthesis I’ and ‘Glycolysis I’ were enriched in genes with decreased expression in lens epithelium but increased expression in lens fiber, indicating their distinct roles in epithelium and fibers during lens morphogenesis.
3.3. Classification analysis of genes based on their expression patterns
To test our hypothesis that co-regulated genes encode proteins that function within specific cell signaling pathways, we examined genes of individual clusters identified in Fig. 3A. Using ToppGenes (Chen et al., 2007) and IPA, we examined the functions of each cluster for enriched GO terms, mouse phenotypes, and signaling pathways. GO term analysis highlighted two fiber clusters, cluster 4 (622 genes) and 17 (492 genes), which showed unique enrichment in the GO term “oxidation-reduction process” (Fig. 11B). Notably, the fiber cluster 4 is the only cluster showing enrichment in the “autophagy” GO term (Fig. 11B). Combining GO terms and mouse phenotypes, we found 3 interesting clusters, the epithelium cluster 13 (96 genes), fiber cluster 11 (58 genes) and 17 (492 genes), as they were enriched in the GO terms of “lens development in camera-type eye” and “abnormal lens morphology” in mouse phenotype. We also noticed that the epithelium-specific clusters 13 (96 genes) and 23 (51 genes) were enriched in “ruptured lens capsule” and “anterior polar cataracts”. In addition, the fiber clusters 11 and 17 were enriched in the “nuclear cataracts” and “abnormal lens fiber morphology”. The epithelium cluster 23 (+1, +1, +1, see Materials and methods), fiber cluster 11 (+1, +1, +1), and fiber cluster 17 (+1, +1, 0) genes showed significantly and continually increased expression in lens epithelium or fiber during lens development (Fig. 3A).
Fig. 3. Functional annotations for tDEG clusters.
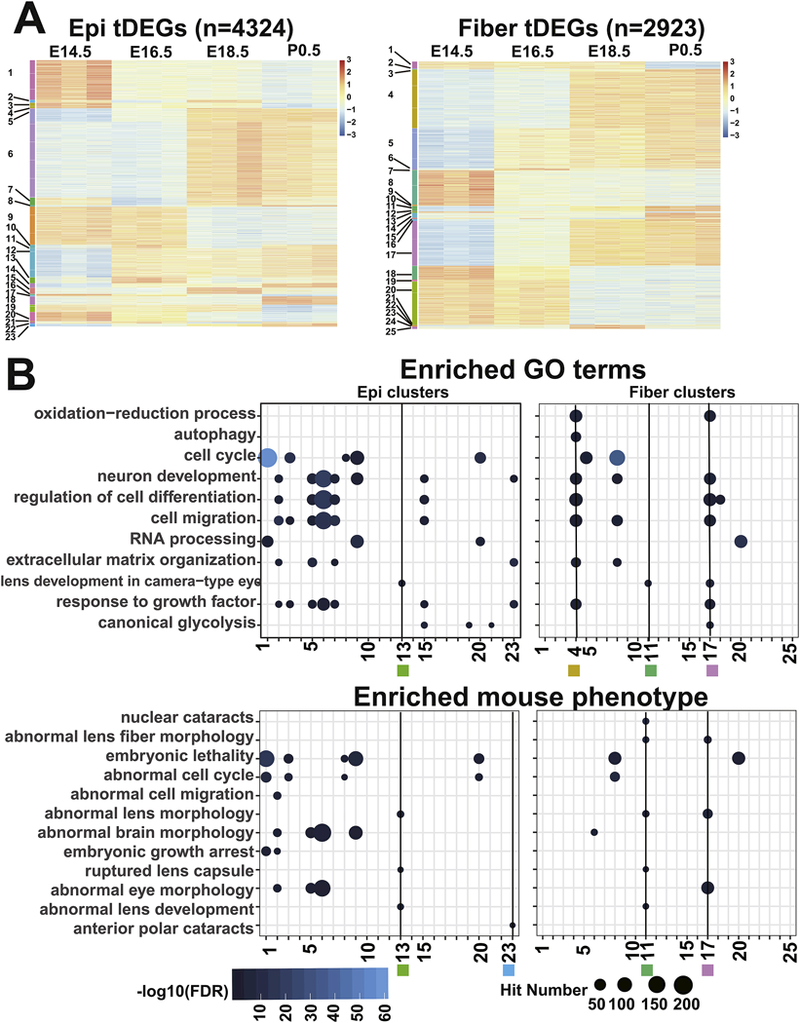
A. The heatmap shows the expression patterns of the 23 epithelium clusters and 25 fiber clusters. Each row represents a gene. The color indicates scaled normalized RNA-seq read counts. B. The dot plot shows enriched GO terms and mouse phenotypes for individual clusters. The terms were first divided into fiber and epithelium enriched and then ranked by their adjusted p values. The blue color corresponds to -log10(FDR) and the dot size corresponds to the number of genes in the GO terms or mouse phenotypes. Black lines with labeled numbers and squares indicate the highlighted clusters.
To explore the roles of canonical signaling pathways in lens differentiation (Cvekl and Zhang, 2017), we performed IPA signaling pathway analysis and found that the epithelium cluster 9 (620 genes), and both the fiber clusters 4 (622 genes) and 17 (492 genes) were highly enriched in “Regulation of eIF4 and p70S6K Signaling”, “EIF2 Signaling” and “mTOR signaling”(Fig. 4A). The epithelium cluster 9 (0, −1, 0) genes showed higher expression in earlier lens development E14.5, E16.5 compared to E18.5 and P0.5 stages. In contrast, the fiber clusters 4 (0, +1, 0) and 17 (+1, +1, 0) genes had higher expression in E18.5 and P0.5 than E14.5, E16.5 (Fig. 4B). These data suggest that specific processes are activated during the initial (E14.5-E16.5) stages of nascent lens epithelium differentiation. Likewise, the expression of genes within the fiber clusters 4 and 17 indicates that fiber cell differentiation- maturation processes occur through the entire time window E14.5-P0.5 analyzed here (Fig. 4B). The increasing trend of expression from E16 to E18 in the fiber cluster 4 (including Eif3k, and Bnip3) was validated by RT-qPCR as shown in Fig. 2. These findings suggest that more novel processes can be learnt from additional in-depth analyses.
Fig. 4. Selected tDEG clusters and enriched signaling pathways.
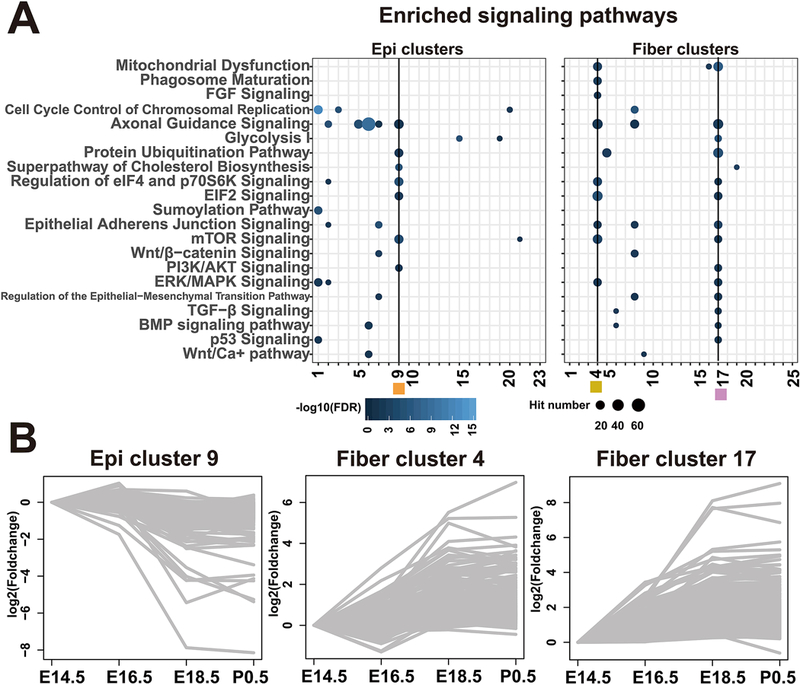
A. Enriched signaling pathways for each temporal cluster, annotated as Fig. 3B. B. The expression profiles of genes in epithelium cluster 9, fiber cluster 4 and fiber cluster 17. Each line represents a gene. The log2(FoldChange) was calculated against the mean FPKM of E14.5.
3.4. Novel mTOR signaling co-expression networks in lens epithelium and fiber
Mtor (differential expression validated in Fig. 2) belongs to a family of phosphatidylinositol kinase-related kinases that regulate metabolism and cell growth (Saxton and Sabatini, 2017). To further elaborate on processes both up-and down-stream of Mtor, we focused here on protein translation, mTOR signaling, autophagy, and mitophagy. Although mTOR signaling was shown to play multiple roles in lens epithelial cell proliferation and migration (Boswell et al., 2017; Qin et al., 2017; Zhang et al., 2016) and rapamycin was reported to induce age onset cataracts in mouse (Wilkinson et al., 2012), the molecular players in lens have not been clearly described, even in previous RNA-seq analyses (Hoang et al., 2014; Khan et al., 2015). Here, we found that mTOR signaling was enriched in genes expressed higher in lens fibers (Fig. 1C). In the temporal gene clusters, we further identified that the epithelium cluster 9 and the fiber clusters 4 and 17 were enriched in mTOR signaling (Fig. 4A). Among these three clusters enriched in mTOR signaling, only the fiber cluster 4 showed additional enrichment of closely related terms, including “Phagosome Maturation” and “FGF signaling”.
When we used the STRING database to define the function interactions of genes in the temporal clusters, including protein-protein interactions and indirect functional associations, we found that in epithelium, mTOR was linked to “mRNA surveillance”, “Ribosome”, “PI3K Akt signaling”, and “Steroid biosynthesis” (Fig. 5A), while in fiber the mTOR signaling was linked to “Oxidative phosphorylation”, “PI3K Akt signaling”, “Ribosome”, “Protein processing in ER”, “proteasome” and “Hippo signaling” (Fig. 5B and Fig. 5C), suggesting distinct functions of these pathways in the two lens compartments.
Fig. 5. Functional interaction of mTOR signaling with genes in the selected tDEG clusters.
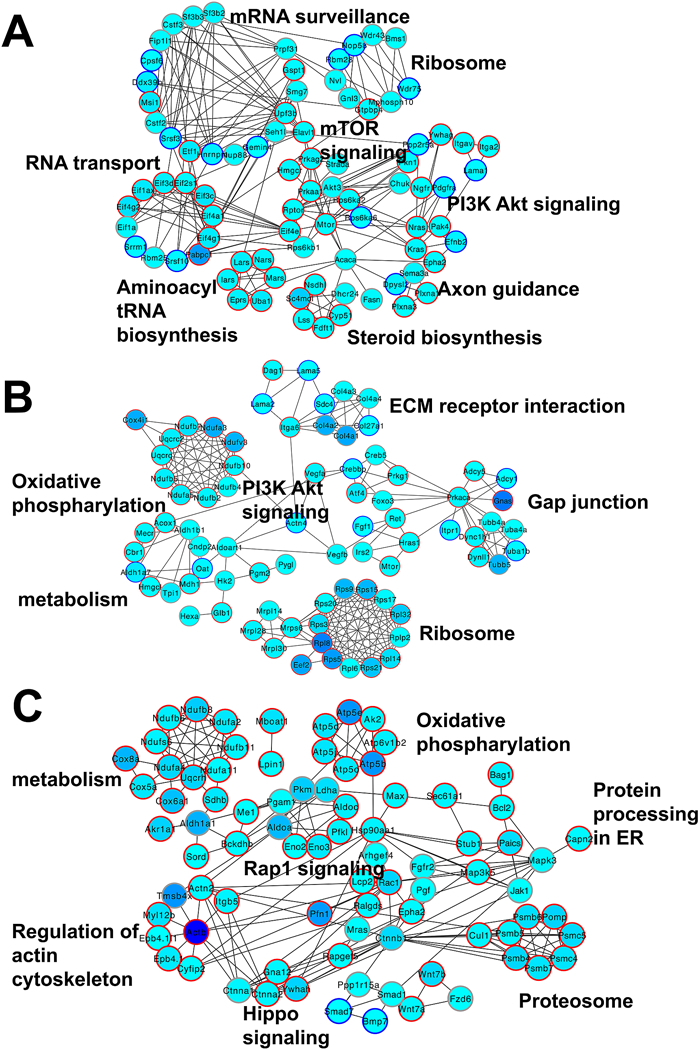
A. The epithelium cluster 9 genes link mTOR signaling to 7 pathways. B. The fiber cell cluster 4 genes link Mtor to 6 pathways. C. The fiber cluster 17 genes show interactions of 7 signaling pathways. The filling color indicates the maximum FPKM of the genes of all time points in either epithelium or fiber. The red circle indicates fiber higher expressed genes. The blue circle indicates epithelium higher expressed genes. The grey circle indicates nondifferentially expressed genes.
Autophagy remains a poorly understood biological process within the lens fibers (Boya et al., 2016; Morishita and Mizushima, 2016). Our data revealed that the activation of this process in lens fiber cells starts from E16.5, consistent with the pattern seen in the fiber cluster 4 (0, +1, 0) (Fig. 3A). mTOR signaling controlled by MAPKs has been previously reported to regulate autophagy in chick lens fibers (Basu et al., 2014). The inhibition of mTORC1 induces premature autophagy in lens fiber cells (Basu et al., 2014). However, the key players connecting mTOR to autophagy in lens remain unclear. We thus collected and examined mTOR signaling-related autophagy genes based on literature report (Fig. 6A). Among all the 30 expressed mTOR signaling-related autophagy genes, only Rptor and Mapk8 showed significantly decreased expression in lens fiber during development, suggesting their possible functions in mTORC1 inhibition (Fig. 6B). Downstream of mTORC1, although the majority of autophagy genes showed significant enrichment in lens fibers, Plekhml was found expressed higher in lens epithelium at E14.5 and only expressed higher in lens fibers from E18.5 to P0.5 (Figs. 2 and 6A). Previous study showed that Plekhm1 worked as a scaffold protein for both homotypic fusion and protein sorting (HOPS)-tethering (HOPS) complex and Atg family proteins (McEwan et al., 2015). Similarly, knockout of Plekhml impeded mTORC1 inhibition that induced autophagy (McEwan et al., 2015). Taken together, Plekhml had spatially distinct expression patterns while Rptor and Mapk8 showed distinct temporal expression patterns in lens development, suggesting they might have distinct functions in modulating autophagy processes in the lens.
Fig. 6. Network analysis for mTOR associated autophagy, translational initiation, and mitophagy.
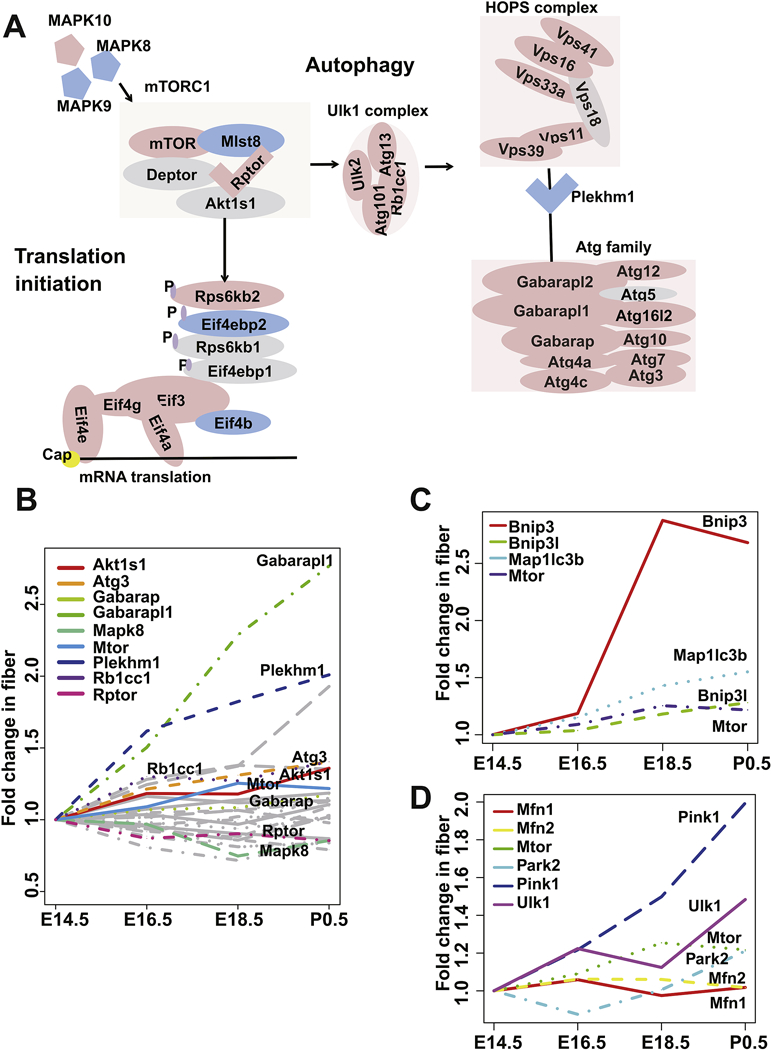
A. mTOR signaling network of autophagy and translational initiation curated from literature. The red color indicates genes with higher expression in fiber at E14.5. The blue color indicates genes with higher expression in epithelium at E14.5. The grey color indicates genes that are non-significantly differentially expressed in epithelium and fiber at E14.5. B. The expression profiles of autophagy genes from mTOR network in fiber development. Each line represents a gene. The grey color indicates genes that are not temporally differentially expressed. The fold change was calculated against the mean FPKM of E14.5. C. The expression profiles of Bnip3 mitophagy signaling associated genes and Mtor in fiber development. D. The expression profile of Park2 mitophagy signaling associated genes and Mtor in fiber development.
Although mTOR-regulated mitophagy was reported in lens, the mechanisms remain to be clarified (Basu et al., 2014). There are two mitophagy pathways: one mediated by Bnip3I/Nix and the other by Parkin. In the fiber cluster 4, we noticed that Mtor was coexpressed with genes in the former pathway, including Bnip3 (Fig. 2), Nix/Bnip3l, and Map1lc3b, while the genes in the latter (Pink1, Park2, Ulk1, Mfn1, and Mfn2) were not temporally differentially expressed in lens fiber (Figs. 6C,D). Together, these results indicate Mtor might regulate mitophagy process in lens through Nix signaling pathway (Brennan et al., 2018).
Interestingly, our data for the first time showed there was a strong enrichment of ‘Translation Initiation’ GO term and ‘Regulation of eiF4 and p70S6K signaling’, ‘EIF2 Signaling’ signaling pathways in genes with higher expression in fiber. Although a recent study has shown that Eif3h regulated translational initiation of crystallin genes in zebrafish (Choudhuri et al., 2013), the potential mechanisms remain unclear in lens. Importantly, mTOR signaling pathway also showed co-enrichment with translational initiation pathways in fiber enriched genes (Fig. 1C), tDEGs with decreased expression from E16.5 to E18.5 in epithelium and increased expression from E14.5 to E18.5 in fiber (Fig. 2S B), lens epithelium cluster 9, fiber cluster 4 and cluster 17 (Fig. 4A). Although Mtor was co-expressed with many Eifs in epithelium cluster 9, only Eif3h, Eif3k (Fig. 2) were found co-expressed with Mtor in the fiber cluster 4. As previously reported in cancer cells (Mamane et al., 2006), Mtor could induce phosphorylation of p70S6K, Eif4ebp1 and cause their dissociation from Eif3 and Eif4e respectively, inducing the translation initiation (Fig. 6A).
Taken together, our analysis for the first time reveals a distinct mTOR signaling related network in lens epithelium and fiber and broadens the roles of mTOR signaling in lens development by uncovering the dynamic spatiotemporal expression patterns of the component genes. Importantly, these studies propose testable hypotheses to examine how Mtor controls protein abundance through modulating translational initiation processes in lens.
3.5. Identification and analysis of novel lens epithelium and fiber specific TFs
The mammalian genome encode ~1,800 TFs that play critical regulatory roles in development and proper function of the organism (Vaquerizas et al., 2009). Lens development is an advantageous system to understand the interplay between signaling pathways and transcription factors (Cvekl and Zhang, 2017); nevertheless, the full account of lens-preferred TFs remains to be established. To address this issue, TFs with expression enrichment in lens versus whole body without the eye was recently explored using the iSyTE 2.0 platform (Kakrana et al., 2018). To extend the study, here we analyzed the TFs in the AnimalTFDB (Zhang et al., 2015) and iSyTE 2.0 database. Among the 1,400 TFs expressed in lens (mean FPKM>1 in any group), 305 TFs (291 listed in iSyTE 2.0) showed higher expression in the epithelium and 135 TFs (123 listed in iSyTE 2.0) higher in lens fibers (Fig. 7). Comparing the lens enrichment in E12.5 embryo (vs whole body) with the fold change of lens epithelium versus fiber in E14.5 lens, we obtained a list of TFs with high expression in lens and significantly differential expression between lens epithelium and lens fiber (23 out of 123 fiber TFs and 12 out of 291 epithelium TFs) (Fig. 7). Specific transcription factors critical to lens development, such as Proxl (Audette et al., 2016), Sox1 (Nishiguchi et al., 1998), Pax6 (Fig. 2) (Sun et al., 2015b; Sun et al., 2016) were lens enriched (FC>8) but they showed relatively small difference (FC<3) between epithelium and fiber. Interestingly, AP-2a (Tfap2a) (Pontoriero et al., 2008) showed a big change (FC>8) between lens epithelium and fiber but it was not so enriched in lens (FC<3). As expected from E14.5, Foxe3 (Medina- Martinez et al., 2005), Hsf4 (Fujimoto et al., 2004), and Maf (Narumi et al., 2014; Ring et al., 2000; Xie et al., 2016) were found to be both lens enriched and differentially expressed between lens epithelium and fiber. Based on the patterns for these known important lens TFs, the present analysis suggests that Dmrta2, Myb, Pbx2, Tulp3 (Fig. 2), and Zfp316 would be excellent novel candidates for regulating early stages of lens formation, while Arid3b, Arntl2, Atf5, Bach2, Carhsp1, Hmg20b, Stat5a (Fig. 2), Stat5b, Tox2, Zfp40, Zfp568, and Zhx2 are predicted to regulate lens fiber cell differentiation (Fig. 7). Stat5a/b would be particularly good candidates to follow up because the JAK-STAT signaling in lens (Ebong et al., 2004; Potts et al., 1998) remains poorly understood and it may also be involved in lens induction (Lleras-Forero et al., 2013).
Fig. 7. TFs with enriched expression in lens epithelium and fiber.
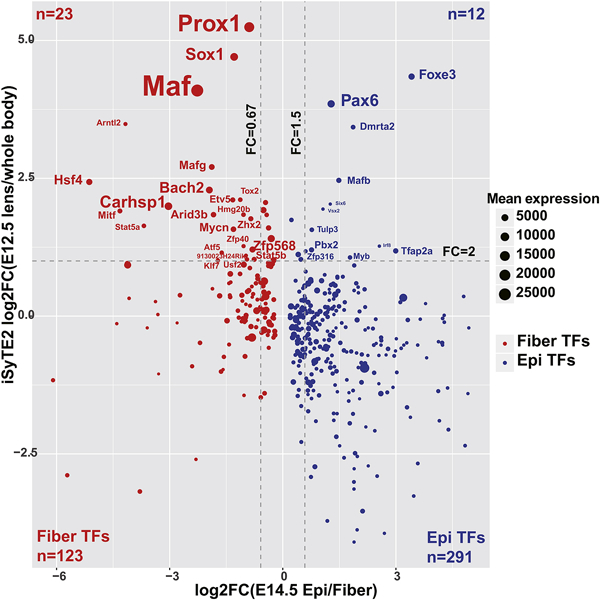
The plot shows lens expressed and spatially differentially expressed transcription factors. Lens enriched (FC>2) and lens epithelium and fiber differentially expressed TFs (FC >1.5 or FC <0.67) are labeled. The blue color indicates E14.5 epithelium higher expressed TFs. The red color indicates E14.5 fiber higher expressed TFs. The dot size corresponds to mean normalized counts in E14.5 lens epithelium or E14.5 lens fiber depending on the enrichment. The text size is proportion to the dot size.
To test the hypothesis that transcription factors with epithelium or fiber-enriched expression can regulate expression of sDEGs, we conducted an unbiased analysis of the promoter regions (−2kb to +2kb of the TSS) of the sDEGs for enriched TF binding motifs, employing Homer software (Heinz et al., 2010). We performed this analysis for the genes with consistently higher expression in lens epithelium (n=3,544, sDEGs up-regulated in lens epithelium) or fiber (n=3,518, sDEGs up-regulated in lens fiber). The analysis revealed a set of common DNA motifs enriched in both groups of genes (Fig. 8). The analysis demonstrated that the E2F motif (rank #1) is most significantly enriched in the group of genes up-regulated in lens epithelium (Fig. 8A). Consistent with these findings, mRNAs encoding E2F family genes (E2f2, E2f3, E2f4, E2f7, and E2f8) were more abundant in epithelium than in lens fibers from E14.5 to P0.5 stages. The E2F family members are critical regulators of G1/S cell cycle progression (Benavente and Dyer, 2015; Hallstrom and Nevins, 2009), so this result is consistent with the sustained proliferative activity within the embryonic lens epithelium. Moreover, earlier functional studies of E2F family members have demonstrated their roles in lens formation (Chen et al., 2000; Chong et al., 2009; Hyde and Griep, 2002; Wenzel et al., 2011)
Fig. 8. Motif analysis for the lens epithelium or fiber higher expressed genes.
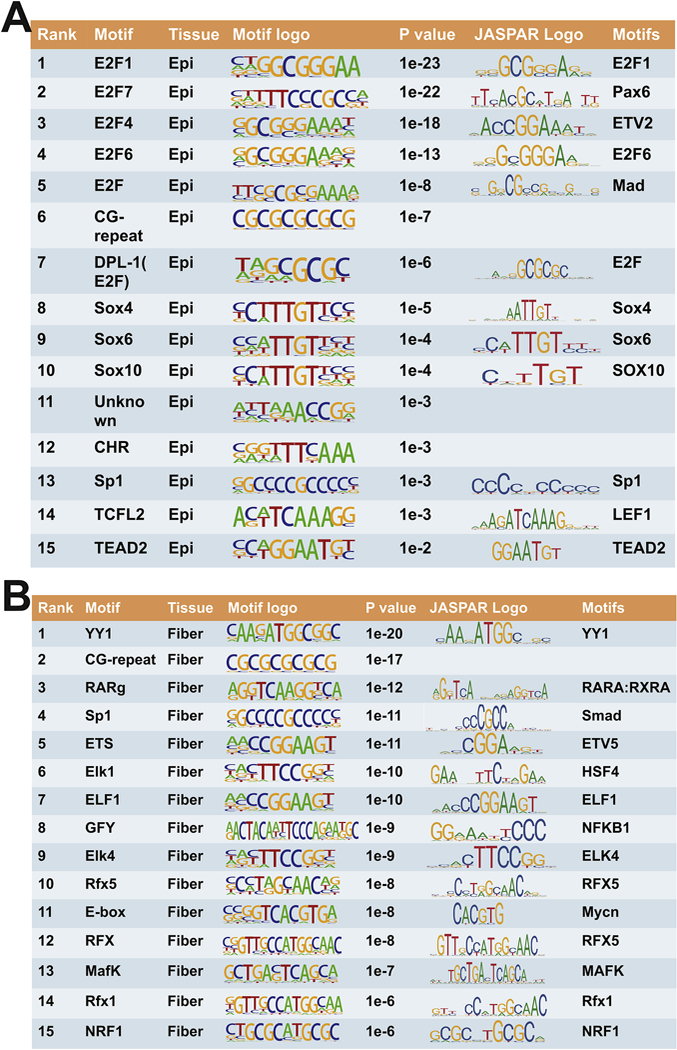
The analysis included 4013 and 3935 of promoter regions (±2kb of the TSSs) from sDEGs in the lens epithelium and lens fibers, respectively.
Additional 14 TF motifs are also worth mentioning (Fig. 8A, rank #2 to 15). The rank #2 sequence is annotated as an E2F7-binding site, but a close inspection found that they resemble Pax6-binding sites (Sun et al., 2015b; Xie and Cvekl, 2011). The sequences ranked #3, 4, 5, 7 are annotated as binding sites for the E2F family; nevertheless motifs #3 and 5 also resemble prototypic Ets and Smad1/5 binding sites, respectively. Motifs #8,9,10 are annotated as Sox-binding sites and Sox1, Sox2, and Sox11 are important regulators of lens differentiation (Cvekl and Zhang, 2017; Kondoh et al., 2004). We propose that motif #11 resembles FoxD1-binding site (not shown, from ISMARA database). Although the binding sites of FoxE3 remain unknown (Blixt et al., 2000; Medina-Martinez et al., 2005), overall similarities between both FoxE3 and FoxD1 DNA-binding domains (Hannenhalli and Kaestner, 2009; Nakagawa et al., 2013) raise the possibility that motif #11 may be recognized by highly abundant FoxE3 in the lens epithelium (Zhao et al., 2018). Motif #13 is annotated as Sp1 -binding site and Sp1 proteins are also studied in lens in context of sumoylation (see below, section 3.7). Motif #14 is annotated as a Tcfl2-binding site and its cognate factor (Tcfl2/Tcf4/Tcf7l2) is a component of Wnt/β-catenin signaling and up-regulates c-Myc expression outside of the lens (Muncan et al., 2006; Shah et al., 2015). Expression of c-Myc is needed for lens epithelial cell proliferation (Cavalheiro et al., 2014). Finally, motif #15 is annotated as TEAD2-binding site and also includes the “core” sequence 5’-GGA(A/T)- 3’ recognized by Ets factors. TEADs are components of Hippo-Yap signaling that also control lens epithelial proliferation (Porazinski et al., 2015; Song et al., 2014; Zhang et al., 2010).
For lens fiber enriched genes, the two most enriched motifs are binding site for YY1 and 10 bp long GC-repeat (Fig. 8B). The YY1 sites and GC-repeats are commonly found in many promoters (Deaton and Bird, 2011; Juven-Gershon and Kadonaga, 2010). Alternatively, the motif #1 weakly resembles Kozak consensus sequence including the ATG triplet (Kozak, 1987), a sequence expected to be frequently found in the +2 kb regions from TSSs. Motif #3 is a direct tandem repeat of the retinoic acid (RA) responsive element (RARE). RA signaling plays multiple roles in lens morphogenesis (Cvekl and Wang, 2009). Interestingly, all three RAR family genes (Rara, Rarb, and Rarg) are abundantly expressed in the lens epithelium and RARβ/RXRβ heterodimers were implicated in transcriptional regulation of the epithelial-preferred aB-crystallin (Gopal-Srivastava et al., 1998). Motif #4 represents Sp1 -binding site and potentially a variant of Smad3-binding site. Motifs #5,6,7,9 are annotated as Ets-binding sites. Etv5 is abundant in lens fibers (Fig. 7) and regulates both Maf and Cryaa gene expression (Xie et al., 2016). In contrast, our data show that additional Etv family genes (Etv1/ER81, Etv3, and Etv6) are preferentially expressed in the lens epithelium. Motif #6 also contains a 5’-TTCNNGAA-3’ heat shock element (HSE) motif that is recognized by lens-specific Hsf4 (Fujimoto et al., 2004; Fujimoto et al., 2008; Somasundaram and Bhat, 2004) and Hsf1 and Hsf2 (Somasundaram and Bhat, 2000, 2004). The 20 bp long motif #8 is annotated as Golgi associated olfactory signaling regulator (Gfy)-binding site;however, this sequence can be also aligned with NF-kB, Proxl, and Etv-binding sites. Motifs #10,12 and 14 are annotated as Rfx5, Rfx, and Rfx1 binding sites. There are no data regarding function of this family of winged helix DNA-binding domain transcription factors in lens. Motif #14 can be also aligned with Smad and Pou2f1/Oct-1 binding sites. Oct-1 regulates lens development and expression of Pax6 (Donner et al., 2007). Motif #11 is a canonical E-box (5’-CANNTG-3’) that can be bound by N-Myc in lens fibers (Cavalheiro et al., 2017) as well as by multiple members of USF family of bHLH transcription factors. USF binds the chicken aA- and δ1 -crystallin promoter and enhancers, respectively (Cvekl et al., 1994; Cvekl et al., 1995). In addition, this sequence can be also aligned with Hif1 a-binding site, 5’- ACGTG(C/G)-3’(Kushida et al., 2016) The Hifla gene is essential for lens morphogenesis (Chen et al., 2008; Shui and Beebe, 2008). Motif #13 is annotated as MafK-binding site and MafG/MafK complex regulates multiple processes including sterol synthesis in the lens (Agrawal et al., 2015). In addition, this site also resembles c-Maf binding site and loss-of- function studies of this transcription factor demonstrated its critical role for crystallin gene expression program (Cvekl and Zhang, 2017). Finally, motif #15 is annotated as the NRF/NKRF-binding site. The NF-kB repressing factor (NKRF) acts as a stress-regulated switch implicated in ribosomal processing and nucleolar homeostasis surveillance (Coccia et al., 2017). It also contains an imperfect E-box (see above). Taken together, analysis of the top 30 enriched motifs at the promoters of sDEGs confirms the known roles of well- characterized TFs in lens development, but also suggests novel ones for future functional and genetic studies.
3.6. Using our RNA-seq data for inferring potential change in epithelium and fiber cells in mouse lens with specific gene knockout
There are a number of opportunities to illustrate how the present RNA-seq data can help understand the functions of genes in lens development. For example, multiple RNA-seq and microarray experiments were conducted to analyze mouse lens with specific gene knockouts, including Pax6 in E14.5 lens (Shaham et al., 2013), Proxi in E13.5 lens (Audette et al., 2016), Mycn in E14.5 lens (Cavalheiro et al., 2017), and Zeb2/Sip1 (Manthey et al., 2014) and b1-integrin/Itgb1 (Wang et al., 2017) in E15.5 lenses. As a range of defects were found in these published studies, we demonstrate here how the present RNA-seq data can be integrated with the published ones to investigate the mechanisms by which inactivation of these regulatory genes could lead to potential abnormal ratios of epithelium to fiber cell population in developing lenses.
Starting from the Prox1 RNA-seq data, we found that 509 of the original DEGs from Prox1 KO vs WT comparison were expressed in our analysis (Fig. 5S). Among them, 179 (35%) were up-regulated and 330 down-regulated in ProxT−1- lenses. Interestingly, we found that the majority (125 genes, 70%) of the up-regulated genes after Prox1 knockout were expressed higher in the E14.5 epithelium, while the majority (311 genes, 94%) of the down- regulated genes were expressed higher in E14.5 fiber cells. This difference is highly significant (p-value < 2.2e-16 by Fisher exact test). Notably, 82% of up- and 67% of down- regulated genes in ProxT−1- lenses were tDEGs in lens development (Fig. 5S). Most of the up- regulated genes were enriched in epithelium clusters 5 and 6, which highlighted the function in neuron development in agreement with Prox1 roles in neural retina (Dyer, 2003) and central nervous system development (Bunk et al., 2016; Karalay et al., 2011). Conversely, lens ProxT1’ down-regulated genes were enriched in fiber clusters 4, 5, 11, 17, and 20 which highlighted RNA processing, RNA stability, cell cycle, oxidative reduction, programmed cell death, mitochondrial electron transport, and lens development. In addition, through inspectionof multiple earlier microarrays and RNA-seq data sets, we found differentially expressed genes of TFs knockout mice versus wild type mice (including Proxl, Mycn, Pax6, and Sip1) were significantly enriched in genes with consistently higher expression in lens epithelium/fiber (Fig. 5S). These findings suggest that the ratios of fiber to epithelium cells in those mutant lenses are significantly different from that of the WT.
To further explore this more quantitatively, we selected 282 lens signature genes that were significantly enriched in either lens epithelium (n=65) or fiber (n=217) (Supplementary File S5, 6). We first tested whether the signature genes can predict percentage of lens epithelium and fiber by simulation (see Materials and Methods). Using Spearman’s correlation to select the best matching samples, we showed that our lens epithelium and fiber RNA-seq samples were most similar to the simulation data with either 100% epithelium or 100% fiber cells, as expected (Fig. 6S). We next tested whether these lens signature genes could be used to infer the percentage of lens epithelium and fiber in the whole lens expression data published previously (Khan et al., 2015). The result indicated that data from both E15 and E18 whole lenses had the highest correlation with simulation data composed of 50–60% of lens epithelium, while data from P1, P3, P6 and P9 whole lens had the highest correlation with simulation data with 40% of lens epithelium (Fig. 6S). These findings support that our simulation methods can be used to predict the ratio of lens epithelium to fiber cells in whole lenses that are used to collect gene expression data.
We thus applied this method to evaluate the cell population difference between the gene knockout lens and the wild type lens. For the Proxl’1’ data, the percentage of lens epithelium detected was changed from 40% to 90% following Proxl inactivation (Fig. 9A), fully consistent with the observed phenotype that Proxl’1’ lens only had a vesicle without any elongated fiber cells (Audette et al., 2016). While in the Mycn’1’ lens, we found that the percentage of epithelium was increased from 40% to 60% in Mycn’1’ lenses (Fig. 9B). Similarly, we found a change of epithelium from 40% to 50% and from 40% to 30% in Sip1 and βΐ-integrin knockout lenses (Fig. 9 C, D), respectively. These results indicate that applications of our lens epithelium and fiber RNA-seq data set can help predict the percentages of lens epithelial and fiber cells from whole lens RNA-seq samples, reveal differentiation defects and thus provide insight into the effect of gene knockout in mouse lens development especially when mutated lenses are grossly malformed.
Fig. 9. Cell population changes in embryonic mouse lens after gene knock out.
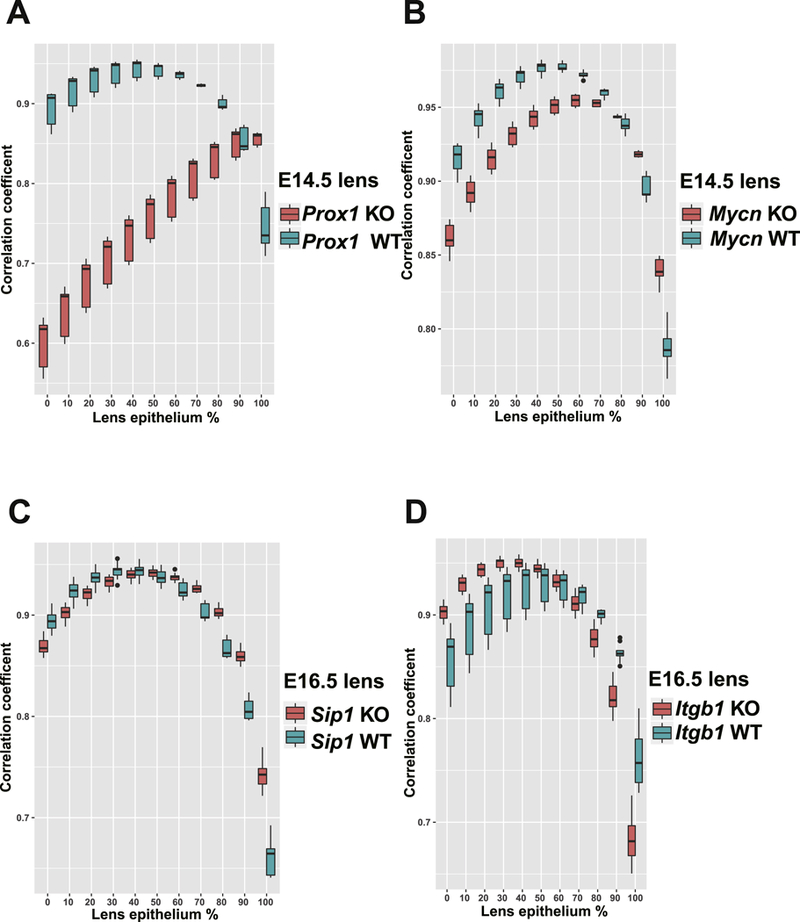
Theboxplot represents Spearman’s correlation coefficient calculated between the simulation data with different percentage of epithelium and real (mixture) data. The red color indicates mutant lens and the cyan color indicates wild type lens. A-D, data for Proxl, Mycn, Sip1 and Itgbl knock out whole lens studies.
3.7. Exploration of sumoylation as a possible mechanism of transcription independent activation of specific transcription factors in lens development
A number of TFs are regulated by sumoylation (Rosonina et al., 2017). We found that the expression of sumoylation pathway in lens was both spatially and temporally regulated: spatially enriched in lens epithelium (Fig. 1A), temporally enriched in the tDEGs with decreased expression in lens epithelium from E14.5 to E16.5, (Fig. 2S), and specifically enriched in the epithelium cluster 1 (Fig. 4A). Consistent with earlier reports (Gong et al., 2014), Sumol mRNA expression was detected in both epithelium and fibers (Fig. 2), while Sumo3 was expressed higher in lens epithelium throughout lens development (Fig. 2, Fig. 10A). Surprisingly, Sumo2 mRNA was very low in lens (Fig. 10B). Although the Sumo1/3 and sumoylation conjugating enzyme E2 Ube2i were stably expressed throughout development, the expression levels of the sumoylation activating enzymes, Sae1, Uba2, were decreased in lens epithelium (Fig. 2, Fig. 10B), supporting the previous finding that both Sumo1 and Sumo3 conjugates were decreased in lens development (Gong et al., 2014).
Fig. 10. Sumoylation model in lens development.
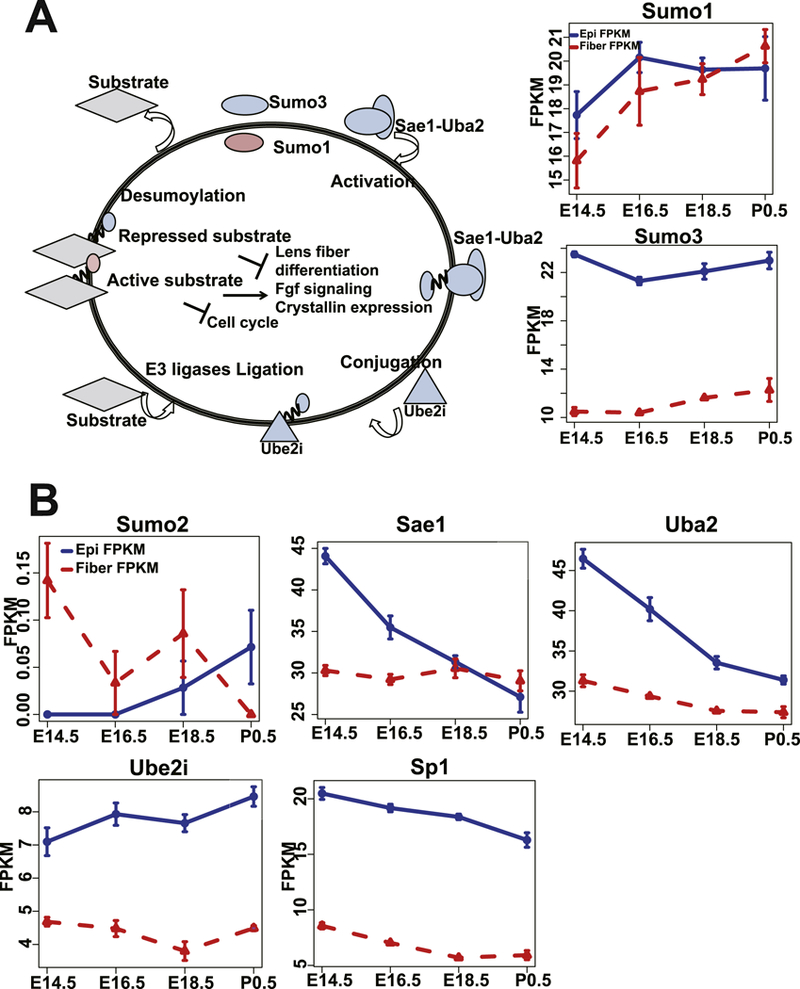
A. The model of sumoylation pathways in lens development and Sumol, Sumo3 expression profiles in lens epithelium and fiber development. The red color indicates Sumo1. The blue color indicates epithelium higher expressed genes at E14.5. B. The expression profiles of Sumo2, Sae1, Uba2, Ube2i and Sp1 in lens development.
Sumoylation of Sp1 and Pax6 were reported previously (Gong et al., 2014; Yan et al., 2010) and both of these proteins are among the most abundant TFs in the lens epithelium (Zhao et al., 2018). Interestingly, we observed an enrichment of Sp1 motif genes up regulated in lens fiber, but Sp1 mRNA was more abundant in epithelium (Fig. 10B). This may be explained by the possibility that Sumo1 modification leads to activation of Sp1 in lens fiber while Sumo3 modification causes its inhibition in lens epithelium. Interestingly, Pax6 Sumo1 sumoylation caused activation of Pax6 in lens (Yan et al., 2010). Additionally, sumoylation of both c-Maf and Prox1 were reported (Li et al., 2012) in lens and c-Jun and Fos sumoylation (Bossis et al., 2005) were also reported in Hela cells. Based on these findings, we propose a model of sumoylation mechanisms in lens development (Fig. 10A), in which Sumo3 modification in lens epithelium mediates the inhibitory roles of some TFs, while Sumo1 modification in both lens epithelium and fiber induces the activation of TFs to regulate embryonic lens development.
3.8. Identification of lens ncRNAs and their potential roles in regulating adjacent genes
ncRNAs represent pleiotropic regulators of gene expression, including subnuclear organization, enhancer activation, post-transcriptional regulation, and allosteric regulation of proteins (Geisler and Coller, 2013). Systematic analysis of ncRNAs and their roles during lens development remains in infancy (Hoang et al., 2014; Wolf et al., 2013). Here we found a total of 675 (5.3% of 12,611) ncRNAs expressed in the mouse lens (Fig. 11A). Among them, 75 were expressed significantly and consistently higher in epithelium and 112 higher in lens fibers. Some of the most expressed long ncRNAs (lncRNAs) in lens were enriched in cataract tissue obtained from age-related cataracts patients, including Tug1 (Li et al., 2017), and Miat (Shen et al., 2016) (Fig. 11B). The functions of these highly expressed ncRNAs, such as Firre, Rmrp, Sox2ot (negative regulator of Sox2, (Amaral et al., 2009; Messemaker et al., 2018)), Neat1 (lncNeat1 binds active chromatin sites, (West et al., 2014), and is regulated by p53 (Adriaens et al., 2016)), Gas5, Six3os1 (regulatory ncRNA of Six3, (Rapicavoli et al., 2011),Rmst (regulates neurogenesis together with Sox2, (Ng et al., 2013), and Gm5607 (Soxlot, co-expressed with Sox1), remain elusive.
Fig. 11. Non-coding RNAs expressed in lens and their co-expressed genes.
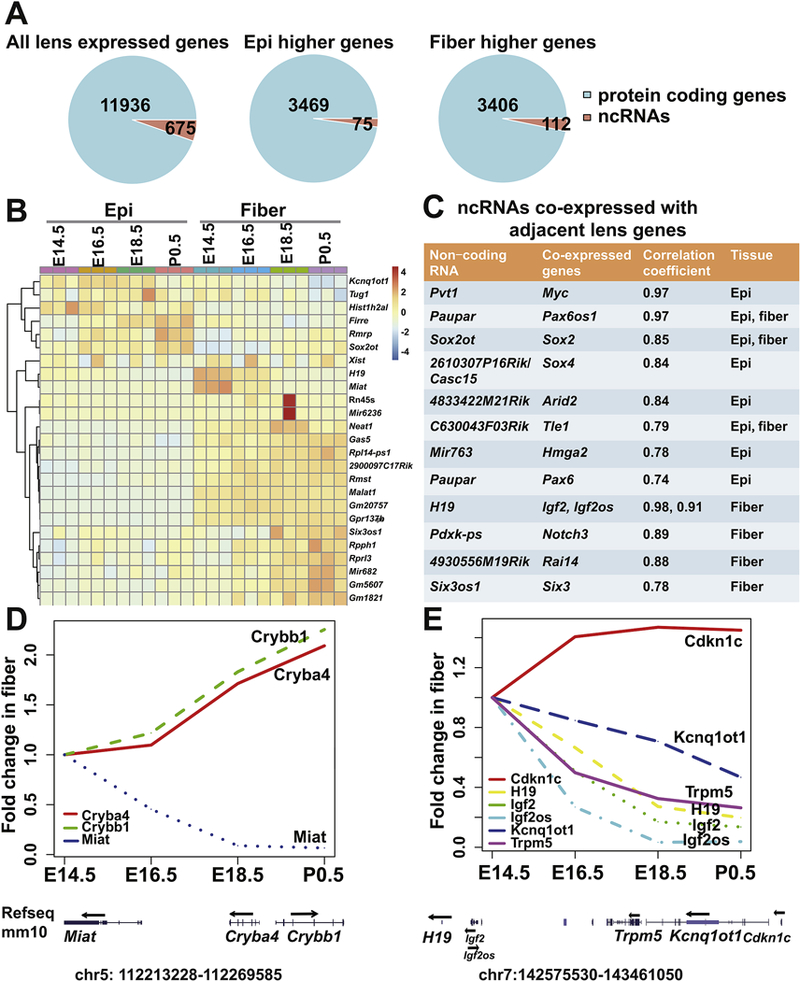
A. The piecharts of ncRNAs for all lens expressed, significantly and consistently higher expressed in either epithelium or fiber. B. The heatmap of the expression of the top 25 ncRNAs. The color indicates scaled normalized RNA-seq read counts. C. Pairs of coexpressed ncRNAs and their adjacent lens genes. D. The expression profiles of Miat, Crybb1 and Cryba4 in fiber development. The locations of the genes in mm10 are also shown. E. The expression profiles of Cdkn1c, H19, Igf2, Igf2os, Kcnq1ot1 and Trpm5 in fiber development.
It was reported previously that many ncRNAs regulated nearby targets on the chromosome, such as Paupar lncRNA and Pax6 (Vance et al., 2014) and Sox2ot lncRNA and Sox2 (Messemaker et al., 2018). Here, we found 220 (128 in epithelium and 92 in fiber) temporally differentially expressed ncRNAs during lens development. Among them, 95 were significantly and positively co-expressed with its nearby genes (correlation coefficient >0.7 and padj <0.05), including transcription factors, lens structural genes and other non-coding genes, such as Sox2ot with Sox2, 2610307P16Rik/CASC15 with Sox4, and Paupar with Pax6 (Fig. 11C). Interestingly, we also noticed that the majority of such pairs of ncRNA (62 out of 95) and its nearby genes were co-expressed in a spatial specific manner during lens development. For example, Paupar lncRNA was only co-expressed with Pax6 in lens epithelium, while Six3os1 lncRNA and Six3 were only co-expressed in lens fiber cells. In contrast to the positive co-expression pattern, we also found 48 pairs of negatively coexpressed genes (r<−0.7 and padj <0.05). Notably, both Cryba4 and Crybb1 mRNAs were significantly and negatively co-expressed with the adjacent Miat lncRNA (padj <0.01). Miat was reported previously as an age-onset cataract enriched lncRNA and could modulate the proliferation, apoptosis and migration of human lens epithelial cells (Shen et al., 2016). Also, Miat-Cryba4-Crybb1 duplication has been detected in patients with congenital cataracts (Siggs et al., 2017). We found that Miat lncRNA was particularly higher expressed in lens fiber cells (Fig. 7S). Temporally, Miat was detected as the only non-coding RNA with decreasing expression in fiber cells as they mature (Fig. 7S). In contrast to down regulation of Miat, the expression of adjacent Cryba4 and Crybb1 continually increased from E14.5 to P0.5 (Fig. 11D).
We also found a group of ncRNAs located within a 1 Mbp region at the mouse chromosome 7 that showed positive co-expression pattern in lens fiber development, including H19, Igf2os and Kcnqlotl, all within the fiber cell cluster 18 (Fig. 11E). In human, this region corresponds to an imprinting region located at 11p1.55, which is related to the Beckwith-Wiedemann Syndrome with CDKN1C/p57 mutations (Li et al., 2001). Interestingly, we found that Cdkn1c mRNA was significantly and negatively correlated with Kcnq1ot1(r=- 0.7), Trpm5 (r=−0.92), H19 (r=−0.85), Igf2 (r=−0.93), Igf2os (r=−0.95) (Fig. 11E) in lens fibers, indicating this methylated region might regulate cell cycle exit in lens development through Cdkn1c (Mancini-Dinardo et al., 2006). In summary, our study found multiple pairs of positively or negatively co-expressed ncRNAs and their adjacent genes during lens development (Supplementary Tables S7 and S8).
4. Conclusion
In this study, we generated and analyzed comprehensive mouse lens RNA-seq data sets that covered E14.5 to P0.5 stages of nascent lens epithelium and fiber cell differentiation, maturation, and remodeling. We have identified abundant transcripts encoding TFs and various components of the lens translational apparatus and critical signaling. These data are compared to proteomic data in our accompanying paper. Analysis of the regulatory regions of sDEGs at E14.5 revealed significant enrichments of E2F, Pax6, Ets, Smad, Sox, Sp1, and other TF binding sites in the lens epithelium. Likewise, TF motifs recognized by RAR/RXR, Sp1, Etv5, Hsf4, N-Myc, and Mafs (c-Maf and MafK) were discovered in genes consistently higher expressed in fiber. In the future, we will perform experiments to analyze the “open” chromatin regions in E14.5 and P0.5 lens epithelium and fibers to better determine the promoters and distal enhancers of the spatially regulated genes. The present data also serves as valuable resources for more follow up studies, such as gene loss-of-function studies in the mouse lens, and studies of lens formation in other model organisms and humans.
Supplementary Material
Highlights.
New RNA-seq datasets to analyze maturation of nascent lens epithelium and lens fibers
Identification of coordinated differentiation processes in mouse embryonic lenses
Resource to study gene regulatory networks that govern lens morphogenesis
Unbiased data analyses confirm and reveal novel lens signaling pathways
Data source for comprehensive analysis for gene loss-of-function mouse lens studies
Acknowledgement
Funding: NIH R01 EY014237 (AC). We thank Dr. Shahina Maqbool, Dr. Shijun Mi and Yongmei Zhao for their extensive advice in RNA-seq data acquisition, the Einstein Epigenomics Shared Facility and high performance computing core for helps. We also thank Yizhou Zhu and Dr. Jian Sun for their advice and help with the RNA-seq library construction.
Abbreviations:
- elF
Elongation initiation factor
- TFs
Transcription factors
- FDR
False discovery rate
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads
- GO
gene ontology
- sDEGs
spatially differentially expressed genes
- IPA
Ingenuity Pathway Analysis
- tDEGs
temporally differentially expressed genes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, Leucci E, Lapouge G, Beck B, van den Oord J, Nakagawa S, Hirose T, Sablina AA, Lambrechts D, Aerts S, Blanpain C, Marine JC, 2016. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nature medicine 22, 861–868. [DOI] [PubMed] [Google Scholar]
- Agrawal SA, Anand D, Siddam AD, Kakrana A, Dash S, Scheiblin DA, Dang CA, Terrell AM, Waters SM, Singh A, Motohashi H, Yamamoto M, Lachke SA, 2015. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non- crystallin genes associated with cataract. Hum Genet 134, 717–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS, 2009. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA (New York, N.Y.) 15, 2013–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand D, Lachke SA, 2017. Systems biology of lens development: A paradigm for disease gene discovery in the eye. Exp Eye Res 156, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W, 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette DS, Anand D, So T, Rubenstein TB, Lachke SA, Lovicu FJ, Duncan MK, 2016. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development 143, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette DS, Scheiblin DA, Duncan MK, 2017. The molecular mechanisms underlying lens fiber elongation. Exp Eye Res 156, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Muraguchi T, Imaoka S, 2013. Role of the hypoxia response pathway in lens formation during embryonic development of Xenopus laevis. FEBS open bio 3, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS, 2009. MEME SUITE: tools for motif discovery and searching. Nucleic acids research 37, W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E, 2014. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res 24, 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, 2009. On the mechanism of organelle degradation in the vertebrate lens. Exp Eye Res 88, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Costello MJ, 2017. The cause and consequence of fiber cell compaction in the vertebrate lens. Exp Eye Res 156, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Sikic H, 2017. The lens growth process. Prog Retin Eye Res 60, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Rajakaruna S, Reyes B, Van Bockstaele E, Menko AS, 2014. Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells. Autophagy 10, 1193–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente CA, Dyer MA, 2015. Genetics and epigenetics of human retinoblastoma. Annual review of pathology 10, 547–562. [DOI] [PubMed] [Google Scholar]
- Beyer EC, Berthoud VM, 2014. Connexin hemichannels in the lens. Frontiers in physiology 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P, 2000. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev 14, 245–254. [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, 2014. Modeling genomic regulatory networks with big data. Trends Genet 30, 182191. [DOI] [PubMed] [Google Scholar]
- Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M, 2005. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Molecular and cellular biology 25, 6964–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell BA, Korol A, West-Mays JA, Musil LS, 2017. Dual function of TGFbeta in lens epithelial cell fate: implications for secondary cataract. Mol Biol Cell 28, 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell BA, Musil LS, 2015. Synergistic interaction between the fibroblast growth factor and bone morphogenetic protein signaling pathways in lens cells. Mol Biol Cell 26, 2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell BA, Overbeek PA, Musil LS, 2008. Essential role of BMPs in FGF-induced secondary lens fiber differentiation. Dev Biol 324, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Esteban-Martinez L, Serrano-Puebla A, Gomez-Sintes R, Villarejo-Zori B, 2016. Autophagy in the eye: Development, degeneration, and aging. Prog Retin Eye Res 55, 206–245. [DOI] [PubMed] [Google Scholar]
- Brennan L, McGreal-Estrada R, Logan C, Cvekl A, Menko S, Kantorow M, 2018. BNIP3L/NIX is required for elimination of mitochondria, endoplasmic reticulum and Golgi apparatus during eye lens organelle-free zone formation. Exp Eye Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Mazot P, Soriano P, 2016. Genetic insights into the mechanisms of Fgf signaling. Genes Dev 30, 751–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunk EC, Ertaylan G, Ortega F, Pavlou MA, Gonzalez Cano L, Stergiopoulos A, Safaiyan S, Vols S, van Cann M, Politis PK, Simons M, Berninger B, Del Sol A, Schwamborn JC, 2016. Prox1 Is Required for Oligodendrocyte Cell Identity in Adult Neural Stem Cells of the Subventricular Zone. Stem cells 34, 2115–2129. [DOI] [PubMed] [Google Scholar]
- Cavalheiro GR, Matos-Rodrigues GE, Gomes AL, Rodrigues PM, Martins RA, 2014. c-Myc regulates cell proliferation during lens development. PloS one 9, e87182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalheiro GR, Matos-Rodrigues GE, Zhao Y, Gomes AL, Anand D, Predes D, de Lima S, Abreu JG, Zheng D, Lachke SA, Cvekl A, Martins RAP, 2017. N-myc regulates growth and fiber cell differentiation in lens development. Dev Biol 429, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauss D, Basu S, Rajakaruna S, Ma Z, Gau V, Anastas S, Brennan LA, Hejtmancik JF, Menko AS, Kantorow M, 2014. Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens. G3 (Bethesda) 4, 1515–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu H, Aronow BJ, Jegga AG, 2007. Improved human disease candidate gene prioritization using mouse phenotype. BMC bioinformatics 8, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Hung FC, Fromm L, Overbeek PA, 2000. Induction of cell cycle entry and cell death in postmitotic lens fiber cells by overexpression of E2F1 or E2F2. Invest Ophthalmol Vis Sci 41, 4223–4231. [PubMed] [Google Scholar]
- Chen Y, Doughman YQ, Gu S, Jarrell A, Aota S, Cvekl A, Watanabe M, Dunwoodie SL, Johnson RS, van Heyningen V, Kleinjan DA, Beebe DC, Yang YC, 2008. Cited2 is required for the proper formation of the hyaloid vasculature and for lens morphogenesis. Development 135, 2939–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L, Fernandez SA, Leone G, 2009. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Molecular and cellular biology 29, 414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri A, Maitra U, Evans T, 2013. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proceedings of the National Academy of Sciences of the United States of America 110, 9818–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RL, Lang RA, 2001. Early eye development in vertebrates. Annual review of cell and developmental biology 17, 255–296. [DOI] [PubMed] [Google Scholar]
- Coccia M, Rossi A, Riccio A, Trotta E, Santoro MG, 2017. Human NF-kappaB repressing factor acts as a stress-regulated switch for ribosomal RNA processing and nucleolar homeostasis surveillance. Proceedings of the National Academy of Sciences of the United States of America 114, 1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Ashery-Padan R, 2014. The cellular and molecular mechanisms of vertebrate lens development. Development 141, 4432–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Sax CM, Bresnick EH, Piatigorsky J, 1994. A complex array of positive and negative elements regulates the chicken alpha A-crystallin gene: involvement of Pax-6, USF, CREB and/or CREM, and AP-1 proteins. Molecular and cellular biology 14, 7363–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Sax CM, Li X, McDermott JB, Piatigorsky J, 1995. Pax-6 and lens-specific transcription of the chicken delta 1-crystallin gene. Proceedings of the National Academy of Sciences of the United States of America 92, 4681–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Wang WL, 2009. Retinoic acid signaling in mammalian eye development. Exp Eye Res 89, 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Zhang X, 2017. Signaling and Gene Regulatory Networks in Mammalian Lens Development. Trends Genet 33, 677–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm R, van Marle J, Quinlan RA, Prescott AR, Vrensen GF, 2011. Homeostasis in the vertebrate lens: mechanisms of solute exchange. Philos Trans R Soc Lond B Biol Sci 366, 12651277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A, 2011. CpG islands and the regulation of transcription. Genes Dev 25, 10101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL, 2007. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol 303, 784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, 2003. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell cycle (Georgetown, Tex.) 2, 350–357. [PubMed] [Google Scholar]
- Ebong S, Yu CR, Carper DA, Chepelinsky AB, Egwuagu CE, 2004. Activation of STAT signaling pathways and induction of suppressors of cytokine signaling (SOCS) proteins in mammalian lens by growth factors. Invest Ophthalmol Vis Sci 45, 872–878. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S, Kato K, Yonemura S, Inouye S, Nakai A, 2004. HSF4 is required for normal cell growth and differentiation during mouse lens development. The EMBO journal 23, 4297–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Oshima K, Shinkawa T, Wang BB, Inouye S, Hayashida N, Takii R, Nakai A, 2008. Analysis of HSF4 binding regions reveals its necessity for gene regulation during development and heat shock response in mouse lenses. The Journal of biological chemistry 283, 29961–29970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura N, 2016. WNT/beta-Catenin Signaling in Vertebrate Eye Development. Frontiers in cell and developmental biology 4, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J, 2013. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestaneh N, Fan J, Fariss RN, Lo WK, Zelenka PS, Chepelinsky AB, 2004. Lens major intrinsic protein (MIP)/aquaporin 0 expression in rat lens epithelia explants requires fibroblast growth factor-induced ERK and JNK signaling. The Journal of biological chemistry 279, 3181331822. [DOI] [PubMed] [Google Scholar]
- Gong L, Ji WK, Hu XH, Hu WF, Tang XC, Huang ZX, Li L, Liu M, Xiang SH, Wu E, Woodward Z, Liu YZ, Nguyen QD, Li DW, 2014. Sumoylation differentially regulates Sp1 to control cell differentiation. Proceedings of the National Academy of Sciences of the United States of America 111, 5574–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal-Srivastava R, Cvekl A, Piatigorsky J, 1998. Involvement of retinoic acid/retinoid receptors in the regulation of murine alphaB-crystallin/small heat shock protein gene expression in the lens. The Journal of biological chemistry 273, 17954–17961. [DOI] [PubMed] [Google Scholar]
- Graw J, 2003. The genetic and molecular basis of congenital eye defects. Nat Rev Genet 4, 876888. [DOI] [PubMed] [Google Scholar]
- Griep AE, 2006. Cell cycle regulation in the developing lens. Semin Cell Dev Biol 17, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Zimmer C, 2011. From cradle to grave: the multiple roles of fibroblast growth factors in neural development. Neuron 71, 574–588. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR, 2009. Balancing the decision of cell proliferation and cell fate. Cell cycle (Georgetown, Tex.) 8, 532–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH, 2009. The evolution of Fox genes and their role in development and disease. Nat Rev Genet 10, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, DeAmicis-Tress C, Cowell TL, Kantorow M, 2005. Identification of global gene expression differences between human lens epithelial and cortical fiber cells reveals specific genes and their associated pathways important for specialized lens cell functions. Mol Vis 11, 274–283. [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, Hejtmancik JF, Horwitz J, Kantorow M, 2004. Identification and functional clustering of global gene expression differences between age-related cataract and clear human lenses and aged human lenses. Exp Eye Res 79, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Limi S, McGreal RS, Xie Q, Brennan LA, Kantorow WL, Kokavec J, Majumdar R, Hou H Jr., Edelmann W, Liu W, Ashery-Padan R, Zavadil J, Kantorow M, Skoultchi AI, Stopka T, Cvekl A, 2016. Chromatin remodeling enzyme Snf2h regulates embryonic lens differentiation and denucleation. Development 143, 1937–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Pirity MK, Wang WL, Wolf L, Chauhan BK, Cveklova K, Tamm ER, Ashery-Padan R, Metzger D, Nakai A, Chambon P, Zavadil J, Cvekl A, 2010. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, 2010. Simple combinations of lineage-determining transcription factors prime cis- regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejtmancik JF, Riazuddin SA, McGreal R, Liu W, Cvekl A, Shiels A, 2015. Lens Biology and Biochemistry. Prog Mol Biol Transl Sci 134, 169–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TV, Kumar PKR, Sutharzan S, Tsonis PA, Liang C, Robinson ML, 2014. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Molecular Vision 20, 27. [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Mivechi NF, 2006. Association and regulation of heat shock transcription factor 4b with both extracellular signal-regulated kinase mitogen-activated protein kinase and dual-specificity tyrosine phosphatase DUSP26. Molecular and cellular biology 26, 3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hyde RK, Griep AE, 2002. Unique roles for E2F1 in the mouse lens in the absence of functional pRB proteins. Invest Ophthalmol Vis Sci 43, 1509–1516. [PubMed] [Google Scholar]
- Jarrin M, Pandit T, Gunhaga L, 2012. A balance of FGF and BMP signals regulates cell cycle exit and Equarin expression in lens cells. Mol Biol Cell 23, 3266–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT, 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana A, Yang A, Anand D, Djordjevic D, Ramachandruni D, Singh A, Huang H, Ho JWK, Lachke SA, 2018. iSyTE 2.0: a database for expression-based gene discovery in the eye. Nucleic acids research 46, D875–d885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalay O, Doberauer K, Vadodaria KC, Knobloch M, Berti L, Miquelajauregui A, Schwark M, Jagasia R, Taketo MM, Tarabykin V, Lie DC, Jessberger S, 2011. Prospero-related homeobox 1 gene (Prox1) is regulated by canonical Wnt signaling and has a stage-specific role in adult hippocampal neurogenesis. Proceedings of the National Academy of Sciences of the United States of America 108, 5807–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Hackett SF, Lee MC, Pourmand N, Talbot CC Jr., Riazuddin SA, 2015. Transcriptome Profiling of Developing Murine Lens Through RNA Sequencing. Invest Ophthalmol Vis Sci 56, 4919–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Uchikawa M, Kamachi Y, 2004. Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int J Dev Biol 48, 819–827. [DOI] [PubMed] [Google Scholar]
- Kong J, Lasko P, 2012. Translational control in cellular and developmental processes. Nat Rev Genet 13, 383–394. [DOI] [PubMed] [Google Scholar]
- Kozak M, 1987. An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic acids research 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushida N, Nomura S, Mimura I, Fujita T, Yamamoto S, Nangaku M, Aburatani H, 2016. Hypoxia-Inducible Factor-1alpha Activates the Transforming Growth Factor-beta/SMAD3 Pathway in Kidney Tubular Epithelial Cells. American journal of nephrology 44, 276–285. [DOI] [PubMed] [Google Scholar]
- Landgren H, Blixt A, Carlsson P, 2008. Persistent FoxE3 expression blocks cytoskeletal remodeling and organelle degradation during lens fiber differentiation. Invest Ophthalmol Vis Sci 49, 4269–4277. [DOI] [PubMed] [Google Scholar]
- Li DW, Gong L, Ji W, Deng M, Liu J, Chen P, Hu W-F, Hu X, 2012. Regulation of Lens Differentiation by Sumoylation with SUMO1/2/3. Investigative Ophthalmology & Visual Science 53, 1305–1305. [Google Scholar]
- Li G, Song H, Chen L, Yang W, Nan K, Lu P, 2017. TUG1 promotes lens epithelial cell apoptosis by regulating miR-421/caspase-3 axis in age-related cataract. Experimental cell research 356, 20–27. [DOI] [PubMed] [Google Scholar]
- Li M, Squire J, Shuman C, Fei YL, Atkin J, Pauli R, Smith A, Nishikawa J, Chitayat D, Weksberg R, 2001. Imprinting status of 11p15 genes in Beckwith-Wiedemann syndrome patients with CDKN1C mutations. Genomics 74, 370–376. [DOI] [PubMed] [Google Scholar]
- Lleras-Forero L, Tambalo M, Christophorou N, Chambers D, Houart C, Streit A, 2013. Neuropeptides: developmental signals in placode progenitor formation. Dev Cell 26, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu FJ, McAvoy JW, 2001. FGF-induced lens cell proliferation and differentiation is dependent on MAPK (ERK1/2) signalling. Development 128, 5075–5084. [DOI] [PubMed] [Google Scholar]
- Lovicu FJ, McAvoy JW, 2005. Growth factor regulation of lens development. Dev Biol 280, 114. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N, 2006. mTOR, translation initiation and cancer. Oncogene 25, 6416–6422. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM, 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20, 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey AL, Lachke SA, FitzGerald PG, Mason RW, Scheiblin DA, McDonald JH, Duncan MK, 2014. Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech Dev 131, 86–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, de Iongh RU, 2010. The lens epithelium in ocular health and disease. The international journal of biochemistry & cell biology 42, 1945–1963. [DOI] [PubMed] [Google Scholar]
- Mathias RT, White TW, Gong X, 2010. Lens gap junctions in growth, differentiation, and homeostasis. Physiological reviews 90, 179–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvoy JW, Dawes LJ, Sugiyama Y, Lovicu FJ, 2017. Intrinsic and extrinsic regulatory mechanisms are required to form and maintain a lens of the correct size and shape. Exp Eye Res 156, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I, 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57, 39–54. [DOI] [PubMed] [Google Scholar]
- Medina-Martinez O, Brownell I, Amaya-Manzanares F, Hu Q, Behringer RR, Jamrich M,2005. Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice.Molecular and cellular biology 25, 8854–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa R, Tyagi M, Harocopos G, Vollman D, Bassnett S, 2016. Somatic Variants in the Human Lens Epithelium: A Preliminary Assessment. Invest Ophthalmol Vis Sci 57, 4063–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messemaker TC, van Leeuwen SM, van den Berg PR, t Jong AEJ, Palstra RJ, Hoeben RC, Semrau S, Mikkers HMM, 2018. Allele-specific repression of Sox2 through the long non-coding RNA Sox2ot. Sci Rep 8, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Mizushima N, 2016. Autophagy in the lens. Exp Eye Res 144, 22–28. [DOI] [PubMed] [Google Scholar]
- Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, Clarke AR, 2006. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Molecular and cellular biology 26, 8418–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML, 2013. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proceedings of the National Academy of Sciences of the United States of America 110, 12349–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi Y, Nishina S, Tokimitsu M, Aoki Y, Kosaki R, Wakui K, Azuma N, Murata T, Takada F, Fukushima Y, Kosho T, 2014. Identification of a novel missense mutation of MAF in a Japanese family with congenital cataract by whole exome sequencing: a clinical report and review of literature. Am J Med Genet A 164a, 1272–1276. [DOI] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW, 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51, 349–359. [DOI] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V, 1998. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev 12, 776781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J, 1981. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation; research in biological diversity 19, 134–153. [DOI] [PubMed] [Google Scholar]
- Pontoriero GF, Deschamps P, Ashery-Padan R, Wong R, Yang Y, Zavadil J, Cvekl A, Sullivan S, Williams T, West-Mays JA, 2008. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Developmental dynamics : an official publication of the American Association of Anatomists 237, 602–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porazinski S, Wang H, Asaoka Y, Behrndt M, Miyamoto T, Morita H, Hata S, Sasaki T,Krens SFG, Osada Y, Asaka S, Momoi A, Linton S, Miesfeld JB, Link BA, Senga T, Shimizu N, Nagase H, Matsuura S, Bagby S, Kondoh H, Nishina H, Heisenberg CP, Furutani-Seiki M, 2015. YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature 521, 217221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts JD, Kornacker S, Beebe DC, 1998. Activation of the Jak-STAT-signaling pathway in embryonic lens cells. Dev Biol 204, 277–292. [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhu Y, Luo F, Chen C, Chen X, Wu M, 2017. Killing two birds with one stone: dual blockade of integrin and FGF signaling through targeting syndecan-4 in postoperative capsular opacification. Cell death & disease 8, e2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S, 2011. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural development 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS, 2000. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127, 307–317. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Akhter A, Dou Y, Babu J, Sri Theivakadadcham VS, 2017. Regulation of transcription factors by sumoylation. Transcription 8, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B, 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic acids research 32, D91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM, 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Schey KL, Petrova RS, Gletten RB, Donaldson PJ, 2017. The Role of Aquaporins in Ocular Lens Homeostasis. International journal of molecular sciences 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Rennoll SA, Raup-Konsavage WM, Yochum GS, 2015. A dynamic exchange of TCF3 and TCF4 transcription factors controls MYC expression in colorectal cancer cells. Cell cycle (Georgetown, Tex.) 14, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, Xie Q, Cvekl A, Shomron N, Davis N, Keydar-Prizant M, Raviv S, Pasmanik-Chor M, Bell RE, Levy C, Avellino R, Banfi S, Conte I, Ashery-Padan R, 2013. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet 9, e1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dong LF, Zhou RM, Yao J, Song YC, Yang H, Jiang Q, Yan B, 2016. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J Cell Mol Med 20, 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Gu S, Yu XS, White TW, Banks EA, Jiang JX, 2015. Connexin Controls Cell-Cycle Exit and Cell Differentiation by Directly Promoting Cytosolic Localization and Degradation of E3 Ligase Skp2. Dev Cell 35, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui YB, Beebe DC, 2008. Age-dependent control of lens growth by hypoxia. Invest Ophthalmol Vis Sci 49, 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggs OM, Javadiyan S, Sharma S, Souzeau E, Lower KM, Taranath DA, Black J, Pater J, Willoughby JG, Burdon KP, Craig JE, 2017. Partial duplication of the CRYBB1-CRYBA4 locus is associated with autosomal dominant congenital cataract. European journal of human genetics : EJHG 25, 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram T, Bhat SP, 2000. Canonical heat shock element in the alpha B-crystallin gene shows tissue-specific and developmentally controlled interactions with heat shock factor. The Journal of biological chemistry 275, 17154–17159. [DOI] [PubMed] [Google Scholar]
- Somasundaram T, Bhat SP, 2004. Developmentally dictated expression of heat shock factors: exclusive expression of HSF4 in the postnatal lens and its specific interaction with alphaB- crystallin heat shock promoter. The Journal of biological chemistry 279, 44497–44503. [DOI] [PubMed] [Google Scholar]
- Song JY, Park R, Kim JY, Hughes L, Lu L, Kim S, Johnson RL, Cho SH, 2014. Dual function of Yap in the regulation of lens progenitor cells and cellular polarity. Dev Biol 386, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Landsbury A, Dahm R, Liu Y, Zhang Q, Quinlan RA, 2009. Functions of the intermediate filament cytoskeleton in the eye lens. The Journal of clinical investigation 119, 1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon WW, Hariharan M, Snyder MP, 2013. High-throughput sequencing for biology and medicine. Molecular systems biology 9, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Chauss D, Wang P, Kantorow M, Zheng D, Cvekl A, 2015a. Chromatin features, RNA polymerase II and the comparative expression of lens genes encoding crystallins, transcription factors, and autophagy mediators. Molecular Vision 21, 19. [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A, 2015b. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic acids research 43, 6827–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhao Y, McGreal R, Cohen-Tayar Y, Rockowitz S, Wilczek C, Ashery-Padan R, Shechter D, Zheng D, Cvekl A, 2016. Pax6 associates with H3K4-specific histone methyltransferases Mll1, Mll2, and Set1a and regulates H3K4 methylation at promoters and enhancers. Epigenetics Chromatin 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL, 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L, 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R, 2010. Fibroblast growth factor signalling: from development to cancer. Nature reviews. Cancer 10, 116–129. [DOI] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP, 2014. The long non-coding RNA Paupar regulates the expression of both local and distal genes. The EMBO journal 33, 296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM, 2009. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10, 252–263. [DOI] [PubMed] [Google Scholar]
- Wang E, Geng A, Maniar AM, Mui BW, Gong X, 2016. Connexin 50 Regulates Surface Ball- and-Socket Structures and Fiber Cell Organization. Invest Ophthalmol Vis Sci 57, 3039–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Terrell AM, Riggio BA, Anand D, Lachke SA, Duncan MK, 2017. beta1-Integrin Deletion From the Lens Activates Cellular Stress Responses Leading to Apoptosis and Fibrosis. Invest Ophthalmol Vis Sci 58, 3896–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel PL, Chong JL, Saenz-Robles MT, Ferrey A, Hagan JP, Gomez YM, Rajmohan R, Sharma N, Chen HZ, Pipas JM, Robinson ML, Leone G, 2011. Cell proliferation in the absence of E2F1–3. Dev Biol 351, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE, 2014. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 55, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J, 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, Woodward MA, Miller RA, 2012. Rapamycin slows aging in mice. Aging Cell 11, 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L, Gao CS, Gueta K, Xie Q, Chevallier T, Podduturi NR, Sun J, Conte I, Zelenka PS, Ashery-Padan R, Zavadil J, Cvekl A, 2013. Identification and characterization of FGF2- dependent mRNA: microRNA networks during lens fiber cell differentiation. G3 (Bethesda) 3, 2239–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Cvekl A, 2011. The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. The Journal of biological chemistry 286, 43259–43271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, McGreal R, Harris R, Gao CY, Liu W, Reneker LW, Musil LS, Cvekl A, 2016. Regulation of c-Maf and alphaA-Crystallin in Ocular Lens by Fibroblast Growth Factor Signaling. The Journal of biological chemistry 291, 3947–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Gong L, Deng M, Zhang L, Sun S, Liu J, Ma H, Yuan D, Chen PC, Hu X, Liu J, Qin J, Xiao L, Huang XQ, Zhang J, Li DW, 2010. Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proceedings of the National Academy of Sciences of the United States of America 107, 21034–21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenka PS, Gao CY, Saravanamuthu SS, 2009. Preparation and culture of rat lens epithelial explants for studying terminal differentiation. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Jin N, Zhang G, Xi Y, Liu H, 2016. SiRNA Targeting mTOR Effectively Prevents the Proliferation and Migration of Human Lens Epithelial Cells. PloS one 11, e0167349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Liu T, Liu CJ, Song S, Zhang X, Liu W, Jia H, Xue Y, Guo AY, 2015AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic acids research 43, D76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D, 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wilmarth PA, Cheng C, Limi S, Fowler VM, Zheng D, David LL, Cvekl A, 2018. Proteome-transcriptome analysis and proteome remodeling in mouse lens epithelium and fibers. Exp Eye Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


