Abstract
Tuberculosis is a global health dilemma. In 2016, the WHO reported 10.4 million incidences and 1.7 million deaths. The need to develop new treatments for those infected with Mycobacterium tuberculosis (Mtb) has led to many large-scale phenotypic screens and many thousands of new active compounds identified in vitro. However, with limited funding, efforts to discover new active molecules against Mtb needs to be more efficient. Several computational machine learning approaches have been shown to have good enrichment and hit rates. We have curated small molecule Mtb data and developed new models with a total of 18,886 molecules with activity cut offs of 10 µM, 1 µM and 100 nM. These datasets were used to evaluate different machine learning methods (including deep learning) and metrics and generate predictions for additional molecules published in 2017. One Mtb model, a combined in vitro and in vivo data Bayesian model at a 100 nM activity yielded the following metrics for 5-fold cross validation: Accuracy = 0.88, Precision = 0.22, Recall = 0.91, Specificity = 0.88, Kappa = 0.31, and MCC = 0.41. We have also curated an evaluation set (n = 153 compounds) published in 2017 and when used to test our model it showed the comparable statistics (Accuracy = 0.83, Precision = 0.27, Recall = 1.00, Specificity = 0.81, Kappa = 0.36, and MCC = 0.47). We have also compared these models with additional machine learning algorithms showing Bayesian machine learning models constructed with literature Mtb data generated by different labs generally were equivalent to or outperformed Deep Neural Networks with external test sets. Finally, we have also compared our training and test sets to show they were suitably diverse and different in order to represent useful evaluation sets. Such Mtb machine learning models could help prioritize compounds for testing in vitro and in vivo.
Keywords: Deep Learning, Drug Discovery, Machine learning, Support Vector Machine, Tuberculosis
Graphical abstract
INTRODUCTION
Mycobacterium tuberculosis (Mtb) infection is causative of tuberculosis (TB), which in 2015 claimed the lives of 1.7 million people with 10.4 million infections reported 1. TB continues to be the focus of intense international efforts to develop new therapeutics to address resistance to first- and second-line drugs 2. The discovery of new TB drug candidates with novel mechanisms of action and a shortened length of treatment is of fundamental importance. Much of the effort has resorted to large high-throughput screens in academia, industry and efforts funded by the NIH and the Bill and Melinda Gates Foundation 3–5. However, the translation of in vitro active compounds coming from these screens and moving them in vivo is fraught with difficulty in terms of finding molecules that balance activity versus good physicochemical and pharmacokinetic properties. For close to a decade we have focused our research on using machine learning models for in vitro Mtb datasets inspired by early Bayesian modeling by Prathipati et al., 6 and our own work using this approach for ADME/Tox properties 7, 8. This has led to modeling of the large Mtb datasets that were made available by SRI/NIAID 9, 10 and smaller datasets from Novartis 11. These models were initially used to score and filter similarly large numbers of molecules identified with pharmacophore methods before eventual testing in vitro 12. This led to three independent prospective tests of models built with in vitro data. One study used Bayesian models to suggest seven compounds for Mtb testing, out of which five were active 13. A second validation of this approach tested 550 molecules and identified 124 actives 14. A third example with an independent group tested 48 compounds and 11 were classed as active 15. A retrospective validation used a set of 1,924 molecules as a test with the various Bayesian models to show the levels of enrichment which in some cases was greater than ten-fold 16. We have since also assessed combining Mtb data sets and then testing the models on data published by other groups17, 18. These combined efforts suggested the importance of having multiple Mtb models and also suggested molecular features important for actives18. The molecular features identified from earlier Bayesian models have been used recently to create new β-lactam antibacterials with activity against Mtb 19. We have additionally developed machine learning models for individual Mtb targets which have been used in a mobile app TB Mobile 20, 21 as well as for drug discovery efforts for ThyX 22 and Topoisomerase I 23. A major hurdle to progressing molecules into the clinic is identifying compounds that have activity in the mouse models of Mtb infection 24. We therefore published a comprehensive assessment of over 70 years of literature resulting in modeling of 773 compounds reported to modulate Mtb infection in mice 24 and more recently updated this with 60 additional molecules 25. Traditionally in vitro and in vivo data and resultant machine learning models have been kept separate and used for different applications.
We have taken the opportunity to curate some recent data from 2015–2016 to update the Mtb models and explore their utility. In addition, we have recently focused on developing software capable of sharing machine learning models based on open source software 26–28 as well as assessing a broader array of machine learning methods including Deep Learning 29, 30. Our goal is to curate additional Mtb data, validate our models while continuing to show their value and predictive potential. Making such TB models accessible could have significant value so that other labs can use them for scoring compounds before testing in global TB drug discovery efforts.
EXPERIMENTAL SECTION
Computing
Most of the computational work was performed using an iMac 21.5-inch iMac with Retina 4K display, 3 GHz Intel core i5, 8 GB Memory 2400 MHz DDR4,1TB HDD, Radeon Pro 555 2048 MB, Magic Mouse 2, Mac OS Sierra (10.12.4). All other computing was done on a single dual-processor, quad-core (Intel E5640) server running CentOS 7 with 96 GB memory and two Tesla K20c GPU. The following software modules were installed: nltk 3.2.2, scikit-learn 0.18.1, Python 3.5.2, Anaconda 4.2.0 (64-bit), Keras 1.2.1, Tensorflow 0.12.1, Jupyter Notebook 4.3.1.
Data Curation
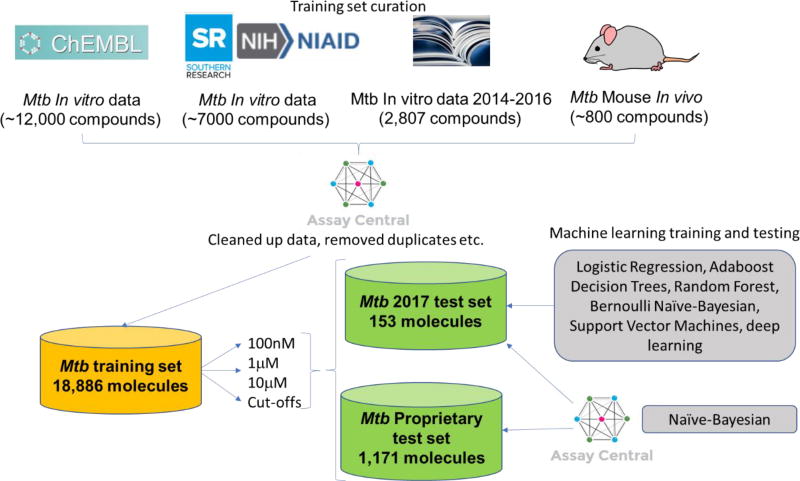
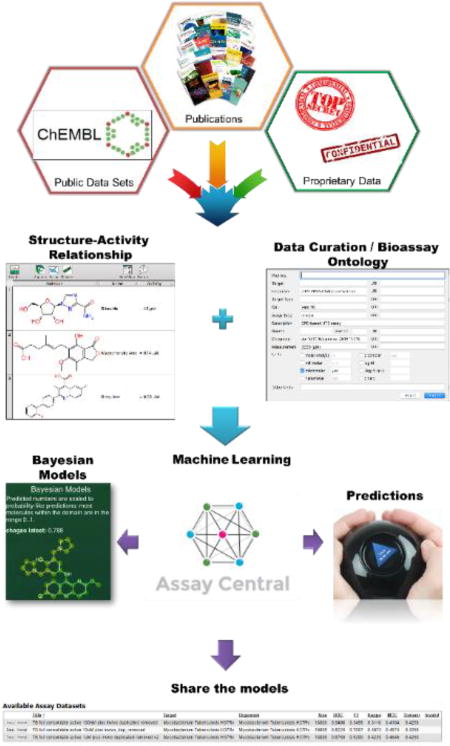
The Mtb data was compiled from four distinct sources. The initial data set was data from approximately 140 primary-literature sources, published between 2014 and 2016, that had been extracted and compiled into a comprehensive list manually. The individual molecular structures of about 2,807 different compounds was created in a format conducive for model building and key data describing the inhibition potency of each of these molecules towards the Mtb H37Ra and H37Rv strain growth were compiled. The minimum inhibition data that was compiled was MIC (minimum inhibition concentration) and were recorded in units of either µg/mL, µM, or nM. Notations were made in each of the files regarding the specific MIC measured (e.g. MIC90, MIC, MIC99, etc.). While these values are slightly different, they are assumed similar enough for model building purposes to be considered approximately the same. If other data was available, such as inhibition % based on a fixed drug concentration, it was recorded in additional columns to be potentially used at a later date. In the cases where there were multiple molecules, a script was run to remove all salts and or all other molecules except for the molecule with the highest molecular weight. These data were extensively curated to guarantee the correctness of the compounds. A second data set that was compiled was extracted from ChEMBL31, 32. A search was performed on the ChEMBL website 32, 33 for a comprehensive list of 1,816 functional assays in the current ChEMBL database involving the target Mtb H37Rv strain. If a data set contained MIC values it was examined and extracted into a usable DataSheet XML (.ds) file format 34 from the latest ChEMBL database using a proprietary script 35,36. The data was checked for consistency between the extracted data and the data on the website. If available, the source of the data was briefly examined to see if a specific target was identified or if it was being tested with knowledge of the target. Independently, a script extracted all of the molecules and the inhibition data from ChEMBL that matched the following criterion: those that included both the parameters Mtb or Mtb H37Rv strain and MIC values. Note this extraction included those that specifically had MIC values only and did not include values of MIC90, MIC99, etc. All of these data were concatenated, and when there were duplicates the MICs were averaged, and then the units were converted to a standard unit of -logM. This combined dataset contained approximately 12,000 compounds. The third data set was compiled from a publicly available SRI data set that contained about 7,000 molecules (a compilation of four published datasets and focusing on dose response data with cytotoxicity16, 37–39). This set was manually curated into a form that was usable to build a model for Assay Central, utilizing the same criteria described above. It should be noted that a script corrected the structure for inappropriately neutrally charged nitro groups. The SRI/NIAID data measured the IC50 or IC90 for growth inhibition, and not the MIC. For the purposes of this model IC50/90 and MIC were estimated to be equivalent. Finally, the model was expanded to include in vivo data which generally assumes compounds would have activity in vitro (~800 compounds) 24, 25. When a conflict existed between what is considered active in the in vivo or in vitro model, the in vivo model trumped was used preferentially. This decision was based on the generality that molecules that demonstrate in vivo efficacy of at least a 1 log10 reduction in lung CFUs are suitably active in vitro. The one notable exception is pyrazinamide, due to a complex mechanism of action that is still the subject of debate 40–48. The In vivo data activity designation did not change based on the 3 cutoffs used in this study. An overview of this data curation process is described in a simplified form (Figure 1).
Figure 1.
Schematic overview of training set curation, test sets and machine learning methods used in this work.
Model Building with Assay Central
We applied best-of-breed methodology for checking and correcting structure-activity data 49 which errs on the side of caution for problems with non-obvious solutions, so that we could manually identify problems and either apply patches, or data source-specific automated corrections. As more data sources are added from diverse sources, the structure processing becomes increasingly important, as standard representation and de-duplication is essential for high quality machine learning model building. The primary deliverable of Assay Central is a “build system” that can routinely recreate a single model-ready combined dataset for each target-activity group. Most software engineering projects of any size use a source code management system such as git, which allows programmers to track the history of each file, resolve conflicts between concurrent branches, and keep their work stored in a safe and secure place while retaining the freedom to work on a local copy. We have repurposed these best practices to the collection of diverse structure-activity datasets, which turns out to be a very similar workflow to collaborative programming: molecular “source files” are edited by team members; our project configuration consists of instructions for gathering the data into categories related by disease target; our compilers filter, clean and merge incoming data-points; and our deliverable is a collection of models that can be used to predict the efficacy and/or toxicity of future compounds of interest, to decide whether they are worth testing in the laboratory. The use of the git source code management program in particular is strategic, since it was designed to manage the sprawling Linux kernel codebase, with thousands of engineers spread across the globe. We have established that the techniques that have been found to be useful for computer programming are similarly effective for cheminformatics data, and our team has been able to perform seamlessly by working on their own local repositories, and periodically pushing their local copy back to the central repository. While many software services exist for explicitly managing SAR data, we began with the presumption that organizing our data as a collection of files in a tree of directories, mixed with build rules - much like computer software source code - would be as effective as using databases (SQL/NoSQL) and custom designed services. The increases in computing power make the relatively ad hoc nature of the system quite sustainable, e.g. the lack of precomputed indexes imposes a considerable computational burden, but this turns out to be quite manageable using off the shelf hardware today. We have included in Assay Central several traditional measurements of model performance, including recall, specificity, precision, F1-Score, Receiver Operating Characteristic (ROC) curve and the area under the curve (AUC), Cohen’s Kappa 50, 51, and the Matthews correlation 52. In some cases, we manually specified a cut-off threshold for active versus. inactive, but we can also use default settings for an automated algorithm which we have described previously 27.
In past studies we have validated and described the generation of Laplacian-corrected Bayesian classifier models 13–15. In this study, three different Bayesian models were created using the combination of 2,807 compounds extracted from the primary literature listed above from 2015–2016, the large data set extracted from ChEMBL of approximately 12,000 molecules, and the SRI/NIAID data set containing ~7,000 9, 10, 13–15, and the ~800 24, 25 compounds from in vivo data as described earlier. The concatenated model, after duplicates were removed, is comprised of 18,886 molecules. The number of actives varied by the activity threshold with values of 645, 2,351, and 7,762 for 100 nM, 1 µM or 10 µM models, respectively. Prior to the model being created each value was converted from µg/mL, µM, or nM into a consistent -logM. A binary threshold of 5, 6, and 7 was then implemented into each model, which is the equivalent to a 10 µM, 1 µM, and 100 nM activity cutoff, respectively. As stated above, the threshold for the in vivo data was set by the experimenters and not by our chosen threshold. The data was either already binary (no action required) or it was converted to binary by applying a threshold (as described above). Each model was internally validated by a five-fold cross-validation, where 20% of the dataset is left out and used as a test set five times. Each 20% left out is randomly removed, but it still retains the representative ratio of the actives:inactives of the total set. Receiver operator characteristic (ROC) plots were generated and the ROC area under the curve (AUC), other model statistics were also calculated in Assay Central. The ROC plots were used to evaluate the predictive ability of the models.
Model Validation
To test the predictive ability of our Mtb models using extended connectivity fingerprints of maximum diameter 6 (ECFP6) we compiled a test set of 153 compounds not in the training set. These data were compiled from ten recently published papers in 2017 53–62 that quantified the growth inhibition of small molecules against Mtb. These were picked essentially at random from PubMed, with the only criterion being that the primary article contained at least one fairly active compound (MIC ≤ 10 µM) and that these data were not available in the current version of ChEMBL 32.
Datasets and Descriptors for Machine Learning Comparison
The Mtb models were used with different types of descriptors in the following comparison of machine learning algorithms. ECFP6 and FCFP6 fingerprints with 1,024 bins were computed from SDF files using RDKit (http://www.rdkit.org/), MACCS keys using 166 substructure definitions were calculated with RDKit Descriptors including 196 2- and 3-dimensional topological, compositional, or electrotopological state descriptors. Toxprint used a set of publicly available set of chemical substructures relevant to toxicity from ChemoTyper (https://chemotyper.org/).
Machine learning comparison
Two general prediction pipelines were developed. The first pipeline was solely built using classic Machine Learning (CML) methods, such as Bernoulli Naïve Bayes, Linear Logistic Regression, AdaBoost Decision Tree, Random Forest, and Support Vector Machine. Open source Scikit-learn (http://scikit-learn.org/stable/, CPU for training and prediction) ML python library was used for building, tuning, and validating all CML models included in this pipeline. The second pipeline was built using Deep Neural Networks (DNN) learning models of different complexity using Keras (https://keras.io/), a deep learning library, and Tensorflow (www.tensorflow.org, GPU training and CPU for prediction) as a backend. The developed pipeline consists of a random splitting of the input dataset into training (80%) and validation (20%) datasets, while maintaining equal proportions of active to inactive class ratios in each split (stratified splitting). All of the models’ tuning and hyper parameters search were conducted solely through five-fold cross validation on training data for better model generalization. Where possible, models were trained using the full unbalanced dataset, i.e., unequal active and inactive compounds. For Support Vector Machines and Linear Logistic Regression using a threshold of 100 nM the models failed to converge and reach a solution. This is likely due to the low number of active compounds (645) compared to inactives (18,241). For these models, 25% of the inactive class was therefore randomly down sampled to create an active, inactive ratio closer to that for the 1 µM threshold dataset.
Bernoulli Naive Bayes
Naïve Bayes method is a supervised learning algorithm based on applying Bayes’ theorem with the “naïve” assumption of independence between every pair of features. Bernoulli Naïve Bayes implements the naïve Bayes training and classification algorithms for data that is distributed according to multivariate Bernoulli distributions; i.e., there may be multiple features but each one is assumed to be a binary-valued (Bernoulli, Boolean) variable. Naïve Bayes learners and classifiers can be extremely fast compared to more sophisticated methods and have been widely used 63. Our Bernoulli Naïve Bayes (BNB) models were tuned and trained using the BernoulliNB class from the Naïve Bayes module of Scikit-learn. Isotonic calibration is a classifier optimization strategy that aims to calibrate the probability scores for a classifier. After training, an isotonic function, i.e., monotonically increasing, is learned mapping the probability scores of the classifier to the fraction of true positive cases at that probability score, creating a more reliable probability estimate 64, 65. In this work, we used the CalibratedClassifierCV available in Scikit-learn to tune our BNB estimator, through five-fold stratified cross-validation based on isotonic regression. The cross-validation generator estimates the model parameter on the train portions of cross-validation split for each split, and the calibration is done on the test cross-validation split of the train dataset. Then, these calibrated probabilities predicted for the folds are averaged and used for prediction. AUC, F1-score and other metrics listed in data analysis section were computed using these calibrated probabilities.
Linear Logistic Regression with Regularization
Logistic regression measures the relationship between the categorical dependent variable and one or more independent variables by estimating probabilities using a logistic function, which is the cumulative logistic distribution, thus predicting the probability of particular outcomes. The L2 binominal regularized logistic regression method was used to classify the activities. A stochastic average gradient optimizer was used in the LogisticRegressionCV class from the Linear Module of Scikit-learn. A five-fold stratified cross-validation method was used in grid search of the best regularization parameter (L2 penalties were in logarithmic scale between 1e−5 and 1e−1). The AUC of ROC was used for scoring the classification (maximizing AUC) performance for each fold of balanced classes’ classification task.
AdaBoost Decision Tree
AdaBoost is a type of “Ensemble Learning” where multiple learners are employed to build a stronger learning algorithm by conjugating many weak classifiers. The Decision Tree (DT) was chosen as a base algorithm in our implementation of the AdaBoost method (ABDT). The AdaBoostClassifier class with 100 estimators and 0.9 learning rate from Scikit-learn ensemble module was used. Similarly to Naïve Bayes, the ABDT models were tuned using isotonic calibration for the imbalanced classes with five-fold stratified cross-validation method.
Random Forest
The Random forest (RF) method is another ensemble method, which fits a number of decision tree classifiers on various sub-samples of the dataset and use averaging to improve the predictive accuracy and control over-fitting. The RandomForestClassifier class with maximum depth of tree 5 and balanced classes weights was used to build the model. The five-fold stratified cross-validation grid search was done using 5, 10, 25, and 50 estimators with the AUC of ROC as a scoring function of the estimator.
Support Vector Machine
Support Vector Machine (SVM) is a popular supervised machine learning algorithm mostly used in classification problems with high dimensional spaces 66. The learning of the hyperplane in the SVM algorithm can be done using different kernel functions for the decision function. C SVM classification with libsvm implementation method from Scikit-learn was used (svm.SVC). The five-fold stratified cross-validation grid search using weighted classes was done for 2 kernels (linear, rbf), C (1, 10, 100), and gamma values (1e-2, 1e-3, 1e-4). The parameter C, common to all SVM kernels, trades off misclassification of training examples against simplicity of the decision surface. A low C makes the decision surface smooth, while a high C aims at classifying all training examples correctly. The Gamma value defines how much influence a single training example has. The larger the gamma value is, the closer other examples must be to be affected. Our implementation of SVM automatically finds the best parameters based on these parameters and saves the best SVM model for activity predictions.
Deep Neural Networks
In recent years, deep artificial neural networks (including convolutional and recurrent networks) have won numerous contests in pattern recognition and machine learning 67–69. This algorithm has also sparked interest in the areas of pharmacology and drug discovery 70 while also stimulating numerous reviews of this still nascent area 29, 71–75. Deep learning addresses the central problem in representation learning by introducing representations that are expressed in terms of other, simpler representations. It is worth noting that a single-layer neural network describes a network with no hidden layers where the input is directly mapped to the output layer. In this work for simplification purposes DNN representation, we counted hidden layers only. Quite often 1–2 hidden layers NN are called shallow neural networks and three or more hidden layers NN, are called deep neural networks.
Two basic approaches to avoid DNN model overfitting are used in training including L2 norm and drop out regularization for all hidden layers. The following hyperparameters optimization was performed using a 3 hidden layers DNN (Keras with Tensorflow backend) and the grid search method from Scikit-learn. The following parameters were optimized prior to final model training: optimization algorithm (SGD, Adam, Nadam), learning rate (0.05, 0.025, 0.01, 0.001), network weight initialization (uniform, lecun_uniform, normal, glorot_normal, he_normal, he_normal), hidden layers activation function (relu, tanh, LeakyReLU, SReLU), output function (softmax, softplus, sigmoid), L2 regularization (0.05, 0.01, 0.005, 0.001, 0.0001), dropout regularization (0.2, 0.3, 0.5, 0.8) and number of nodes all hidden layers (512, 1024, 2048, 4096).
The following hyperparameters were used for further DNN training: SGD, learning rate 0.01 (automatically 10% reduced on plateu of 50 epochs), weight initialization he_normal, hidden layers activation SReLU, output layer function sigmoid, L2 regularization 0.001, dropout is 0.5. The binary cross entropy was used as a loss function. To save training time, an early training termination were implemented by stopping training if no change in loss were observed after 200 epochs. The number of hidden nodes in all hidden layers was set equal to number of input features (number of bins in fingerprints). The DNN model performance was evaluated on up to eight hidden layers DNNs.
Molecular property distribution
AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area were calculated from input SD (structural data) files using Discovery Studio 4.1 (Biovia, San Diego, CA) 24.
Principal components analysis
To assess the applicability domain of the in vitro data for the Mtb training and test sets we used the union of these sets to generate a principal component analysis (PCA) plot based on the interpretable descriptors selected previously (AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area) for machine learning, as well as using ECFP6 fingerprints. In addition, we have also compared the Mtb data with a set of 19,905 antimalarials for comparison of a different biological / chemical property space 76, 77 and used the combination to generate a PCA.
Clustering data
We have previously described the Honeycomb clustering (Molecular Materials Informatics, Inc., Montreal, Canada) 25 which is a greedy layout method for arranging structures on a plane in a meaningful way using a reference compound as the focal point placed on a hexagonal grid pattern. This method uses ECFP6 fingerprints for all similarity comparisons using the Tanimoto coefficient. This approach was used with the complete training and test set for compounds.
Determination of minimum inhibitory concentration (MIC) against M. tuberculosis
Methods previously described by us 78, 79 were used to generate MIC data with the M. tuberculosis H37Rv strain either grown in 7H9 broth (Becton, Dickinson and Company 271310), plus 0.2% glycerol (Sigma G5516), 0.25% Tween 80-20% (Sigma P8074) and 20% 5x ADC or a culture grown in Middlebrook 7H9 supplemented with 10% ADS (albumin-dextrose-sodium chloride), 0.2% glycerol and 0.05% tyloxapol to an OD600 0.7–1. Alamar blue or the resazurin assays were used 80.
Statistical analysis
Means for descriptor values for active and inactive compounds were compared by two tailed t-test with JMP v. 8.0.1 (SAS Institute, Cary, NC). In this study, several traditional measurements of model performance were used, including specificity, recall, precision, F1-Score, accuracy, Receiver Operating Characteristic (ROC) curve and AUC, Cohen’s Kappa 50, 51, and the Matthews correlation 52. For the metric definitions, we will use the following abbreviations: the number of true positives (TP), the number of false positives (FP), the number of true negatives (TN), and the number of false negatives (FN). Specificity or TN rate = TN/ (TN + FP). Model Recall (also known as the True Positive Rate or Sensitivity) can be thought of percentage of true class labels correctly identified by the model as true and is defined: . Similarly, model precision (also known as the Positive Predictive Value) is the probability a predicted true label is indeed true and is defined: . The F1-Score is simply the harmonic mean of the Recall and Precision: . Accuracy is another measure of model robustness, and is the percentage of correctly identified labels out of the entire population: . The ROC curve can be computed by plotting the Recall vs the False Positive Rate (FPR) at various decision thresholds T, where . In this study, all constructed models are capable of assigning a probability estimate of a sample belonging to the true class. Thus, we can construct an ROC curve by measuring the Recall and FPR performance when we considered a sample with a probability estimate > T as being True for various intervals between 0 and 1. The AUC can be constructed from this plot and can be thought of as the ability of the model to separate classes, where 1 denotes perfect separation and 0.5 is random classification. Another measure of overall model classification performance is the Matthew’s Correlation Coefficient (MCC), which is not subject to heavily imbalanced classes and is defined as . As a measure of correlation, its value can be between −1 and 1. Cohen’s Kappa (CK), another metric estimating overall model performance, attempts to leverage the Accuracy by normalizing it to the probability that the classification would agree by chance (pe) and is calculated by , where .
RESULTS
Our previous Mtb machine learning models were primarily based on data generated from high throughput screens produced several years ago by SRI/NIAID 16, 37–39. Therefore, we decided to take advantage of the growing recent additional Mtb data in the literature and curate new datasets with likely increased structural diversity over the older datasets and that utilized different cutoffs for activity.
Model Building and Validation
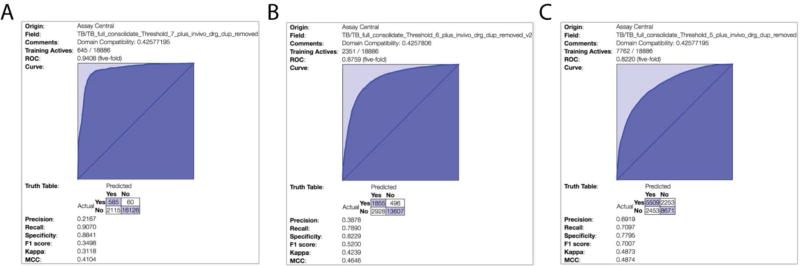
We curated ~2,700 compounds from the primary literature published in 2015–2016, a data set extracted from ChEMBL of approximately 12,000 molecules, the SRI/NIAID data set containing ~7,000 molecules 9, 10, 13–15, and the ~800 24, 25 compounds from in vivo data. After duplicate removal our training set consisted of 18,886 molecules. This was then used to initially build three Bayesian models with different activity thresholds. Assessment of the three Mtb models with different thresholds showed that the ROC AUC values dramatically increased as the activity cut off decreased from 10 to 0.1 µM, with values of 0.82, 0.87, and 0.94, respectively (Supplemental Table 1). The ROC values did not necessarily correlate with the F1-score and MCC (Figure 2, Supplemental Table 1). Our initial model validation performed in Assay Central used the various models to predict the activity of known FDA-approved drugs after removing duplicates present in the training set. This validation was exemplified with our 1 µM activity threshold model, where out of the top 18 scoring compounds 17 were known antibacterials (Supplemental Figure 1) and cyclosporine, which was previously shown to have antitubercular activity 81. The top scoring compound was rifaximin, which has previously been shown to have antitubercular activity 82.
Figure 2.
Tuberculosis model 5-fold cross validation statistics and receiver operator characteristic plots. 5-fold cross validation ROC plots and supplementary statistical analysis of three Mtb models built with 18,886 molecules using Assay Central. A) 100 nM threshold, B) 1 µM threshold, C) 10 µM threshold. ROC AUC and MCC ranges from 0.94-0.82 and 0.41-0.49, respectively.
We additionally curated a set of 153 published in vitro antitubercular compounds from 2017. To assess the differences between the 2017 test set and the Mtb training set we looked at the Tanimoto similarity score distribution (Supplemental Figure 2). Each value represents the ECFP6 fingerprint Tanimoto score between each one of the molecules in the 2017 test set and the most similar compound in the Mtb training set. The average Tanimoto score is 0.35 ± 0.17 (std dev). The histogram shows that the highest occupied bin is 0.3> × >0.2 and that 89.8% of the compounds have a closest Tanimoto similarity score of less than 0.7 (an arbitrary cut off above which one would consider similar compounds). These data show that the Mtb training set and 2017 test set are dissimilar when using ECFP6 fingerprints, the metric used to predict activity.
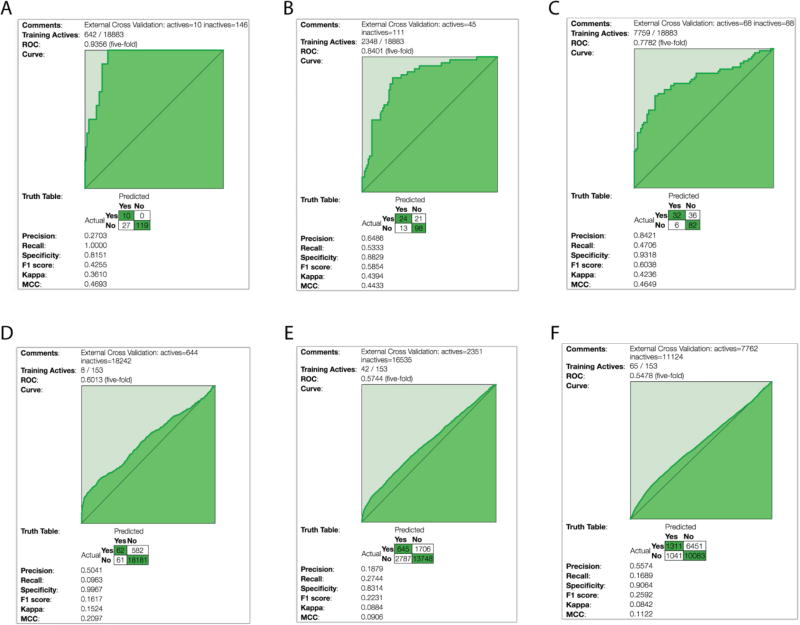
This test set was also used to build a Bayesian model with good internal statistics (Supplemental Figure 3). The three 18,886-compound Mtb models built in Assay Central with different activity cutoffs also demonstrated good external predictive abilities when using the 153 compounds as a test set. The three different 18,886-compound Mtb models, with various activity thresholds, exhibited ROC values of 0.78, 0.84, and 0.93 corresponding to thresholds of 10 µM, 1 µM, and 100 nM, respectively (Supplemental Table 1). Each combination of modeling method and activity cutoff afforded a model with different strengths, with the most relaxed threshold having the best precision and specificity while the most stringent threshold had the foremost recall (Figure 3A–C, Supplemental Table 1). For comparison, we also tested the in vitro data only model. For this validation we removed a large portion of the ChEMBL data and used it as a test set (4,499 molecules), with the remainder of the data as the training set (13,713 compounds). The compounds that were used for the test set were essentially chosen at random. The set consisted of the non-overlapping molecules between the compounds extracted from ChEMBL by organism (Mtb H37Rv, MIC only) and the rest of the molecules in the model. The 1 µM model fared well with a ROC and MCC score of 0.79 and 0.39, respectively, while the 100 nM model predicted moderately with ROC and MCC values of 0.66 and 0.21, respectively. While this is a large validation set and a good test of this ChEMBL model, unfortunately the training set is reduced by approximately 25% weakening its predictive ability (Supplemental Figure 4). These examples above also show the importance of having multiple models at different thresholds.
Figure 3.
External validation ROC plots and supplementary statistical information. Three 18,886 Mtb molecule models A) 100 nM threshold, B) 1 µM threshold, C) 10 µM threshold which were used as a training set with the 153 molecule library used as a test set. D-F) 153 molecule set used as a training set and the larger 18,886 molecule library used as a test set.
Simple descriptor analysis
To assess the chemical property space of the training and test set used we studied the simple interpretable descriptors used to define the chemical property space of compounds namely: AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area. Analyses of these descriptors revealed several surprising differences between the active and inactive molecules in our Mtb models. We internally compared the actives versus inactives in both the 18,886 compound Mtb sets at different cutoffs and for the smaller 153 compound Mtb test set. For the 18,886 compound set we analyzed these differences for all three of our models, each with a different threshold. In all of the larger models there was a statistical difference between four of the eight descriptors (p <0.0001). One of the most significantly different descriptors between data sets is AlogP. At a low threshold (10 µM) the lipophilicity is virtually identical between active and inactive compounds, but as stringency increases there is a trend to decrease the hydrophobicity of the active compounds by 0.3 units. Molecular weight is significantly different in all of the models, increasing in the actives as the threshold increases. This trend is mimicked with hydrogen bond acceptors, with a dramatic ~33% increase with the strictest threshold. These data suggest that both molecular weight and the number of hydrogen bond acceptors are both important criteria for increased antitubercular activity. Similar trends are observed with the smaller 153 compound test dataset, but with less statistical significance (Table 1–4).
Table 1.
Differences in descriptors between active and inactive Mtb compounds - Training set (mean±SEM) 100 nM.
| Number | Class | AlogP | MW | Number of Aromatic Rings |
Hydrogen Bond Acceptors |
Hydrogen Bond Donors |
Number of rings | Rotatable Bonds |
FPSA |
|---|---|---|---|---|---|---|---|---|---|
| 18241 | inactive | 3.80 ± 0.01 | 378.59 ± 1.02 | 2.62 ± 0.008 | 4.51 ± 0.02 | 1.10 ± 0.009 | 3.48 ± 0.01 | 5.35 ± 0.04 | 0.23 ± 0.00072 |
| 645 | active | 3.50 ± 0.074 | 427.54 ± 5.43 | 2.10 ± 0.044 | 6.04 ± 0.10 | 1.43 ± 0.05 | 3.39 ± 0.05 | 6.75 ± 0.19 | 0.25 ± 0.0038 |
| 2 tailed T test p value | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 | 0.09 | <.0001 | <.0001 |
Table 4.
Differences in descriptors between active and inactive Mtb compounds -Test set (mean±SEM) 1 µM.
| Number | Class | AlogP | MW | Number of Aromatic Rings |
Hydrogen Bond Acceptors |
Hydrogen Bond Donors |
Number of rings |
Rotatable Bonds |
FPSA |
|---|---|---|---|---|---|---|---|---|---|
| 111 | inactive | 4.22 ± 0.10 | 396.8 ± 8.84 | 2.34 ± 0.07 | 3.98 ± 0.18 | 0.93 ± 0.07 | 4.33 ± 0.07 | 3.31 ± 0.22 | 0.14 ± 0.005 |
| 42 | active | 5.15 ± 0.16 | 454.69 ± 15.0 | 2.09 ± 0.13 | 5.19 ± 0.29 | 0.62 ± 0.12 | 4.43 ± 0.12 | 4.28 ± 0.38 | 0.16 ± 0.008 |
| 2 tailed T test p value | <.0001 | 0.0008 | 0.09 | 0.0005 | 0.033 | 0.49 | 0.022 | 0.053 |
Model Property Space Analysis and Principal Component Analysis
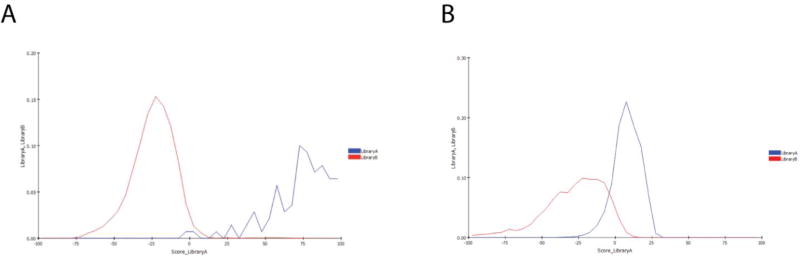
Comparison of the 153 compound test set and the 18,886 compound training set using global fingerprints in Discovery Studio showed a low similarity score of 0.0095 (a value of 1 would equal identity). There were 93,299 total global fingerprint bits, 883 common global fingerprint bits, 677 unique fingerprint bits in the test set and 91,739 unique fingerprint bits in the training set. The similarity score is calculated by creating an independent, global fingerprint for all ligands in both data sets and the comparison metric is the Tanimoto similarity score between the two fingerprints. A histogram shows clear separation of the training set and this independent evaluation set with minimal overlap suggesting the datasets are very different (Figure 4A).
Figure 4.
Histograms of the Bayesian model distances as calculated in Discovery Studio. A) 18,886 molecule Mtb library (Red) vs. 153 compound 2017 test set (blue). B) 18,886 molecule Mtb library (Red) vs. 19,905 anti-malaria set (blue).
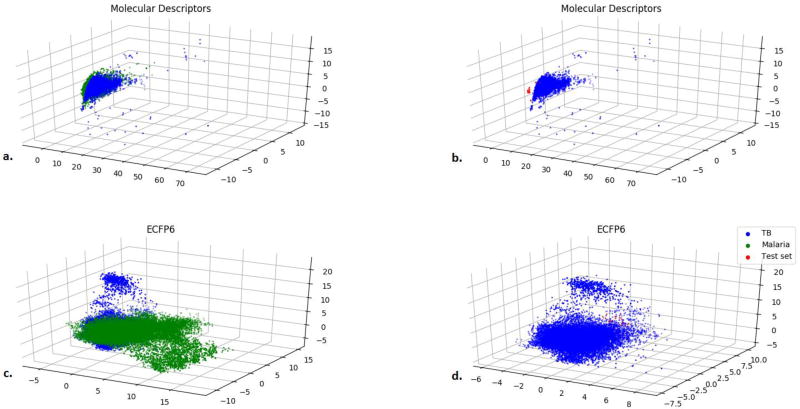
The ability of the 18,886 compound Mtb models to predict the test set even though they are very dissimilar suggests that this Mtb model can successfully predict activity of a novel set of compounds that are far from the training set. To visualize these differences further, we created a Principal Component Analysis (PCA) using ECFP6 fingerprints alone. This PCA showed that the evaluation set is partially overlapping the training set (Figure 5D). This suggests that the applicability domain of the training set is sufficient to predict the activity of new compounds using these descriptors. To expand on this further, we looked at the ability of using the set comprised of the 153 Mtb compounds to predict the larger 18,886 compound Mtb models (the previous training set). The predictions varied from nearly random to weak with ROC scores of 0.55–0.60, dependent on the threshold (Figure 3D–F). These data show that the 18,886 compound Mtb model can predict the smaller 153 compound molecule set, but not vice versa.
Figure 5.
Principal Component Analysis of Mtb datasets using different descriptors and fingerprints. a.) 9 Molecular descriptors for TB Training Set (blue) and Malaria Set (green), b.) 9 Molecular descriptors for TB Training Set (blue) and TB Test Set (red), c.) ECFP6 fingerprints for the TB Training Set (blue) and Malaria Set (green), and d.) ECFP6 fingerprints for TB Training Set (blue) and TB Test Set (red). The TB training set consists of 18,886 Mtb compounds. The TB test set consists of 153 molecules. The malaria training set consists of 19,905 molecules.
As the 18,886 compound Mtb models are generally able to predict the smaller evaluation 153 compound set using ECFP6 fingerprints we wanted to ascertain the chemical property space occupied by each model. A PCA was generated with the following descriptors: AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area. The three components represent 77% of the variance (Figure 5B). The 153 compound test set is shown to be almost completely buried within the 18,886 compound Mtb training set.
To give a sense of scale as to the potential diversity of the 18,886 compound Mtb library we plotted a similar size set of compounds tested as antimalarials (19,905 compounds from multiple laboratories) alongside the Mtb compounds in a PCA using multiple descriptors as described earlier (Figure 5A and 5C). The malaria set nearly comprehensively overlap the Mtb data set using these descriptors, suggesting that they occupy a similar chemical property space. The ECFP6 descriptors alone suggest that the Mtb and malaria set do have some limited overlap when using the compare libraries function in Discovery Studio, with a global fingerprint similarity score of 0.1745. There are a total of 177,031 global fingerprint bits, 30,885 common bits, 84,409 unique bits in the malaria set, and 61,737 unique bits in the Mtb library. Interestingly, using ECFP6 fingerprints the Mtb training set has a much larger diversity than the malaria set (Figure 5B) as calculated from the Bayesian model in Discovery Studio. A PCA between the compounds tested for malaria and Mtb using only the ECFP fingerprints show that these fingerprint descriptors can differentiate these two datasets as the colored areas appear distinct for each set of compounds with the compounds tested for antimalarial activity spanning a larger area in the plot (Figure 5C). Our results also suggest that the results of a PCA analysis to understand the chemical property space covered by 2 datasets is dependent on the type of molecular descriptors used.
Clustering
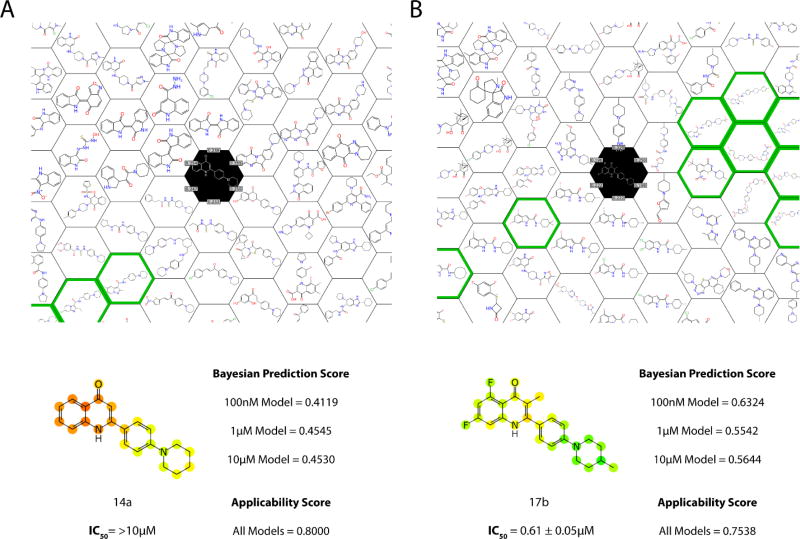
We have previously briefly described Honeycomb clustering 25 and this approach was used in Assay Central to visualize the complete training and select compounds in the 153 compound test set (Figure 6). This can be used as an independent guide to predictions and represents an approach that is different and yet is potentially complimentary to the Bayesian prediction.
Figure 6.
Examples of the scoring output from Assay Central for the test set 60. At the top is a hexplot which shows other molecules in the training set that are similar to our scored compound. The numbers surrounding the compound of interest (black) are Tanimoto scores showing quantitatively the similarity between the nearest neighbors. The green outlined molecules are active in the model chosen (1µM 18,886 Mtb model). The image shows the 2-D molecular structure with each atom colored based on the scores of each of the represented fingerprints. It is scored as a gradient- green is active and red is inactive. The applicability score is the similarity between the molecule of interest and the model.
External Testing of Different Machine Learning Algorithms
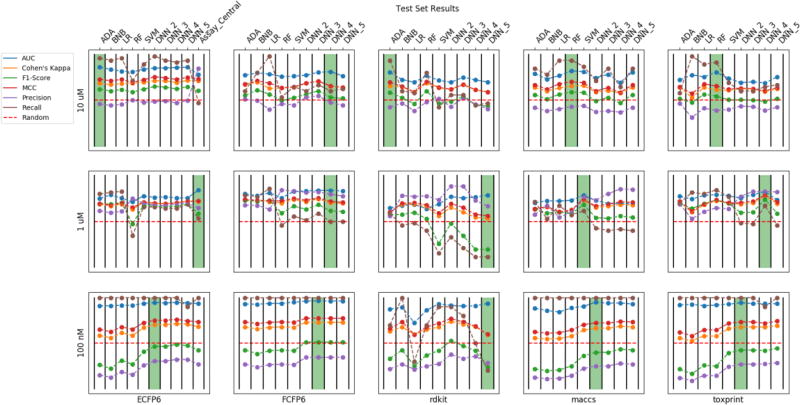
We have also compared different machine learning algorithms that can be used to build models30. We tested the classic machine learning models Logistic Regression, Adaboost Decision Trees, Random Forest, Bernoulli Naïve-Bayesian and Support Vector Machines as well as deep learning algorithms with 2–5 layers using various molecular descriptors. These comparisons were performed with the three separate 18,886 compound Mtb models with threshold cutoffs of either 100 nM, 1 µM or 10 µM. We used this dataset because it is our biggest curated dataset. Comparison as measured by five-fold cross validation, of the machine learning models using the various descriptors suggests that for these Mtb models DNN is the superior algorithm at this cutoff (Supplemental Figure 5). To ensure that these models were not “over-trained”, a separate experiment was done where 20% of the training set (representative of the ratio of active:inactive) was left out and used as a test set. These comparisons show that SVM and DNN outperformed the other machine learning algorithms on accuracy as well as for other statistical analyses for all of the thresholds tested (Supplemental Figure 6). Deep learning algorithms were overall the most efficacious for all the descriptors using all the metrics for training (Supplemental Figure 5) and cross validation (Supplemental Figure 6), including the entire 1 µM threshold set of models and for the 10 µM models: MACCS, RDKit, and toxprint descriptors. However, when using the 153 compound set as an external test set there was more variability observed as to which algorithms performed the best for each descriptor type and based on each metric (Figure 7, Supplemental Tables 2–6). For ease visualization we have highlighted the model that performed the best based on AUC. For example, for the ECFP6 descriptors the ADA algorithm performed best at 10 µM, Assay Central (Bayesian) was best at 1 µM and DNN at 100 nM. These results would suggest a difference between internal testing and external testing.
Figure 7.
Results of various classic machine learning algorithms and deep learning on the 153 compound TB test set using different evaluation metrics. The green bar represents the highest ROC AUC for each category. Machine learning algorithms compared include: Logistic Regression = LR, Adaboost Decision Trees = ADA, Random Forest = RF, Bernoulli Naïve-Bayesian = BNB and Support Vector Machines = SVM, DNN = Deep Neural Network with variable layers. Further details can be found in Supplemental Tables 2–6.
Large Test Set Analysis of Bayesian and DNN Models
Mtb inhibition data was generated in a single laboratory across many antitubercular drug discovery projects for 1,171 proprietary compounds with MIC data. We used our three 18,886 compound Bayesian models generated with Assay Central as well as DNN models to predict the molecules at each activity threshold. We compared the test set molecules to this training set and it had an average closest Tanimoto similarity score of 0.42 ± 0.15. The highest occupied bin is 0.4>x>0.5 and 94.5% of the molecules have a Tanimoto similiarity score of less than 0.7 (Supplemental Figure 7). We found that the non-overlapping 1,171 molecule test set with the Bayesian model at a cutoff of 1 µM performed the best (Supplemental Figure 8A–C) ROC = 0.68, MCC = 0.14, F1 score = 0.24. When the test set is used to predict the training set the statistics degrade (Supplemental Figure 8D–F). This test set was also used to build a model (Supplemental Figure 9) which had excellent statistics ROC = 0.90, MCC = 0.66, F1 score = 0.81. Which will likely be further evaluated outside this study. Interestingly, the DNN models with ECFP6 fingerprints appear to poorly predict this 1,171 compound test set as the ROC values were low across all hidden layers evaluated (Supplemental Figure 10). The 100 nM model appears to show this most clearly with very poor ROC values. The 1 µM DNN model performs the best with MCC and F1 scores slightly lower than obtained for the Bayesian model (Supplemental Table 7).
DISCUSSION
Over the past decade, whole-cell HTS approaches have been used to identify novel compounds with antitubercular activity, resulting in very low hit rates 37–39. Virtual screening using machine learning modeling utilizes data (actives and inactives) from these screens and smaller scale studies to improve the time and cost efficiencies of hit discovery. Machine learning has been similarly used in many areas of biomedical and environmental research 83–88. This study is a natural extension of our prior machine learning studies applied to Mtb 9–11, 13–18 which demonstrated an enhancement in hit rate. To further leverage public data to enhance the performance of our models, we have now curated a large, chemically diverse Mtb dataset through careful analysis of in vitro data mined from various sources, such as the primary literature and ChEMBL, and also included in vivo data. Machine learning models were then constructed with this combined data with the caveat that MIC, MIC90, and IC50 are equivalent values even though they may be several-fold different from each other. This is a limitation to our model, but based on the retained predictive ability of models built with this training set to correctly categorize the external compound test set, this assumption appears not to be detrimental. The initial models were built using the circular topological descriptor extended connectivity fingerprints ECFP6 with Assay Central. Internal validation of these models was achieved by using five-fold cross validation (Figure 2). One of the main purposes in the report is to show the validity of our newly curated 18,886 compound library using an array of machine learning approaches, descriptors and metrics. In addition, we have also created a separate test set of 153 compounds from the antitubercular literature from 2017. Library analysis software and Bayesian statistics with ECFP6 descriptors suggest that these test and training sets are nearly independent of one another. Surprisingly, analysis using PCA with different descriptors suggested that they both occupy the same chemical property space. Using different descriptors profoundly alters the way the chemical property space is defined. This would suggest that reliance on different descriptors and fingerprints can markedly change how 2 sets of compounds are perceived to cover chemical property space.
In the process of this study we have shown that Assay Central using the Bayesian algorithm and ECFP6 descriptors performs generally the best with the 153 compound test set as well as a set of 1171 proprietary compounds tested against Mtb. The 153 compound set represents compounds published in 2017 alone and is a different approach to predicting how a model can be used to prospectively predict ‘newer’ compounds. While others have used methods such as time-split validation 89 this is distinct to our approach.
We have used the calculate diversity metrics protocol in Discovery Studio to assess the inactives, actives, and complete datasets for the 2017 Mtb test set, the proprietary test set, as well as the full Mtb training set at different threshold cutoffs for activity (100 nM, 1 µM, 10 µM). When the average Tanimoto similarity score across these datasets was analyzed it showed a couple of key points: 1) the average for each dataset ranged from 0.10–0.22, suggesting a strong internal diversity within the datasets (Supplemental Figures 11–13) 2) the two smaller datasets showed an apparent increase in similarity in the actives with a more stringent activity cutoff (Supplemental Figures 11–12). This suggests that, for the two test sets, the stricter cutoffs tend to have a reduced ECFP6 diversity. In other words, with higher cutoffs similar fingerprints are selected. This decreased diversity may yield a reduced ability for the stricter cutoff models to predict novel compounds. The larger Mtb training set (Supplemental Figure 13) does not show this trend, suggesting that if the dataset is large enough, this reduced diversity based on the cutoffs analyzed is not relevant. The average Euclidean distance using multiple metrics (ALogP, MW, number of H-bond donors, number of H-bond acceptors, number of rotatable bonds, number of rings, number of aromatic rings, molecular fractional polar surface area) does not show the same trends (Supplemental Figures 14–16). For the two smaller datasets (Supplemental Figures 14–15) the average Euclidean distance is slightly higher in the actives, suggesting these molecules are actually more diverse than their inactive counterparts. There is the opposite trend in the Mtb training set for the 100 nM and 1µM cutoffs (Supplemental Figure 16), with virtually no difference at the 10 µM cutoff. These inconsistencies suggest that there is unlikely to be a difference between the diversity, using these metrics, in the actives versus inactives datasets. One of the most pronounced differences in diversity between the actives versus inactives for the different datasets is the average number of fingerprints per molecule (Supplemental Figures 17–19) and the average number of assemblies per molecule (Supplemental Figures 20–22). Analysis of these metrics show that in the strictest cutoff of 100 nM both of these metrics are ~2–3 fold higher in the actives as compared with the averages found in the entire model. This suggests that those molecules identified as active at 100 nM tend to be larger, more complex molecules. A trend towards diminished molecular complexity with more relaxed cutoffs continues in the majority of the models assessed. This agrees with our data (Tables 1–4), where we see a significant increase in the average molecular weight in the actives versus inactives.
Using Bayesian modeling for testing of the 153 compound test set with ECFP6 descriptors alone we show a range of ROC AUC scores from 0.78–0.93, dependent on the activity threshold. This illustrates the ability of the training set to accurately predict the test set. Interestingly, the lowest threshold (10 µM) had the best precision and specificity, but the worst recall. Additionally, the 100 nM cutoff model had the best recall so it selected all the true positives, but had the lowest precision meaning that it identified many false positives. The Kappa and MCC scores were similar, with the 100 nM cut-off model having the highest MCC and the 1 µM cut-off model having the best Kappa score. We have recently shown that ROC values may be an optimistic measure of a model while Kappa and MCC may be more realistic 30. By using multiple models for Mtb we show that each activity threshold has different strengths and it is possible that all three models could be used when scoring molecules to aid in compound selection. Using Assay Central to build the Bayesian models predicted the external evaluation test set with the additional metric of accuracy showing a score as high as 0.83 (100 nM threshold model). While the 18,886 Mtb set was able to predict the 153 compound Mtb test set, the inverse is not true. This suggests that the larger set has a better predictive ability with ECFP6 descriptors.
We have looked at the effect of dataset size by focusing on the ChEMBL dataset only and leaving out a subset. We found that the larger Mtb model slightly edged out the smaller ChEMBL data set, but the statistics were nearly identical. In a previous study we saw a similar plateauing of model ROC when assessing different dataset sizes 18.
One method to expand the usefulness of these larger models using Bayesian models would be to utilize a nearest neighbors approach. If a model weighted the final prediction score based on both the Bayesian score and the kNN score (weighted by the model size) this may yield more accurate predictions. Our honeycomb clustering approach may partially address this (Figure 6) as a molecule that is close to known active molecules in the training set may provide more confidence in the prediction. This will be further expanded upon by us in future studies with different datasets.
In this study we analyzed the chemical property space of both the Mtb 153 compound test set and the 18,886 compound Mtb training set via PCA (Figure 5). We showed that when looking at an array of descriptors, there is considerable overlap between these sets. It is notable that there are some compounds that lie outside of the cluster containing the majority of the molecules tested against Mtb. A cursory inspection of these outliers suggests that the majority of the molecules are inactive, showing that these were likely novel compounds that were tested against Mtb but did not have any activity. Further, we looked at the differences in the descriptors between the active and inactive molecules for each model built at different thresholds. The greatest differences were found in the 100 nM cut-off model, but the majority of trends were continued. These statistics suggest that active compounds have an increased molecular weight, number of hydrogen bond acceptors, rotatable bonds, and FPSA (polar surface area descriptor). As the model becomes more stringent for activity, the AlogP decreases from 3.80 to 3.50. Previous studies, however, have shown that reliance on individual descriptors for predicting antitubercular activity may be an oversimplification 9, 10. By further exhaustive analysis we have also been able to compare the active and inactive molecules using average Tanimoto similarity with ECFP6 descriptors, average Euclidean Distance with many simple descriptors, average number of fingerprint features and average number of assemblies (Supplemental Figure 11–22) which confirm what we were seeing in Tables 1–4.
To test the predictive ability of different machine learning algorithms we generated cross validation statistics for each method. Beyond just testing each method with ECFP6 descriptors, we also tested multiple, publicly accessible descriptors (Supplemental Figures 5–6). Assay Central currently uses a Näive Bayesian algorithm with ECFP6 descriptors 26, 27, so we were only able to compare the Assay Central Mtb models built with this descriptor for the 153 compound external test set (Figure 7). One of the differences between Assay Central and Bernoulli Naïve-Bayesian would be in the classification of true/false hard outcomes. For the Assay Central implementation, we do an analysis of the ROC and use that to decide on a threshold for the unscaled output. The BNB implementation does it by making 2 predictions: one for positive and one for negative and taking whichever is higher. Comparison of these datasets using different machine learning algorithms, demonstrates deep learning and SVM appear to be superior methods regardless of the descriptor type for training (Supplemental Figure 5) and leave out validation (Supplemental Figure 6) whereas for external testing DNN models did not perform as well (Figure 7, Supplemental Tables 2–6) with Adaboost Decision Trees performing the best at 10 µM, Assay Central at 1 µM and DNN_2 at 100 µM. This was shown most clearly with results for the 1171 compound test set (Supplemental Figure 10). This suggests we may have to optimize the descriptors or model parameters further. These results suggest that DNN may not always perform the best although we report a limited example for a single biological activity based on two external test sets. Further testing is needed to understand if DNN offers benefits for prospective testing for other datasets as it does for cross validation 30.
We were able to use our machine learning models to predict Mtb inhibition data from our laboratory generated over several years. This predictive analysis of a very large dataset of over 1,000 structurally diverse compounds (Supplemental Figure 7) suggests that the Bayesian models perform reasonably well at the 1 µM cutoff for predicting activity (Supplemental Figure 8B). In addition, a separate Bayesian model generated with this dataset suggests we could use this to predict future data from this laboratory that would be more comparable as the data would come from the same laboratory (Supplemental Figure 9).
Additional limitations of this work include that we have focused from the outset on drug sensitive Mtb and not specifically modeled drug resistant Mtb. Our rationale for this is there is far less data in the literature that would enable building machine learning models against drug resistant strains in the same way that we have been able to build models versus the sensitive strain. It is still feasible that a model constructed with drug sensitive data could help us identify molecules active against drug resistant strains.
In conclusion, there is considerable publicly available antitubercular growth inhibition data generated over the last decade. Investing significant effort in data curation has allowed us to build accurate, predictive models for Mtb employing an array of machine learning methods, descriptors, statistical metrics and testing scenarios. The three models which we have focused on with different cutoff values could be used either together or in consensus as described for other Mtb models 18. Future work could certainly explore other descriptors and machine learning algorithms beyond those tested here, while a critical factor is the continual updating of the models with new experimental data that is a rate-limiting step in building accurate Mtb models. Assay Central offers a unique approach to building, updating and sharing models for Mtb that is essential if we are to increase the efficiency of drug discovery. Our future efforts will involve further prospective testing of the different machine learning algorithms with collaborators using data from the same laboratory as well as models derived after data curation across published literature and public databases.
Supplementary Material
Table 2.
Differences in descriptors between active and inactive Mtb compounds - Training set (mean±SEM) 1 µM.
| Number | Class | AlogP | MW | Number of Aromatic Rings |
Hydrogen Bond Acceptors |
Hydrogen Bond Donors |
Number of rings |
Rotatable Bonds |
FPSA |
|---|---|---|---|---|---|---|---|---|---|
| 16535 | inactive | 3.82 ± 0.01 | 377.39 ± 1.07 | 2.62 ± 0.009 | 4.47 ± 0.02 | 1.10 ± 0.01 | 3.47 ± 0.01 | 5.35 ± 0.04 | 0.23 ± 0.00075 |
| 2351 | active | 3.63 ± 0.04 | 400.24 ± 2.84 | 2.51 ± 0.023 | 5.17 ± 0.05 | 1.16 ± 0.03 | 3.53 ± 0.03 | 5.72 ± 0.10 | 0.24 ± 0.002 |
| 2 tailed T test p value | <.0001 | <.0001 | <.0001 | <.0001 | 0.04 | 0.07 | 0.0006 | <.0001 |
Table 3.
Differences in descriptors between active and inactive Mtb compounds - Training set (mean±SEM) 10 µM.
| Number | Class | AlogP | MW | Number of Aromatic Rings |
Hydrogen Bond Acceptors |
Hydrogen Bond Donors |
Number of rings |
Rotatable Bonds |
FPSA |
|---|---|---|---|---|---|---|---|---|---|
| 11124 | inactive | 3.79 ± 0.02 | 371.02 ± 1.31 | 2.56 ± 0.011 | 4.38 ± 0.02 | 1.12 ± 0.01 | 3.41 ± 0.01 | 5.29 ± 0.05 | 0.22 ± 0.00092 |
| 7762 | active | 3.79 ± 0.02 | 393.50 ± 1.56 | 2.66 ± 0.012 | 4.82 ± 0.03 | 1.09 ± 0.01 | 3.58 ± 0.01 | 5.54 ± 0.05 | 0.23 ± 0.00110 |
| 2 tailed T test p value | 0.86 | <.0001 | <.0001 | <.0001 | 0.13 | <.0001 | 0.0006 | <.0001 |
Acknowledgments
Biovia is kindly acknowledged for providing Discovery Studio and Pipeline Pilot to S.E. and J.S.F. Professor Nancy Connell and Dr. Riccardo Russo of Rutgers University and Professor William R. Jacobs, Jr. and Dr. Catherine Vilchèze of the Albert Einstein College of Medicine are kindly acknowledged for generating Mtb growth inhibition data used in the proprietary test set. Dr. Mary A. Lingerfelt is kindly thanked for the table of contents graphics assistance.
Grant information
This work was supported by Award Number 1R43GM122196-01 “Centralized assay datasets for modelling support of small drug discovery organizations” from NIH/ NIGMS. Some of the datasets were previously built partially with funding from 9R44TR000942-02 “Biocomputation across distributed private datasets to enhance drug discovery” from the NIH National Center for Advancing Translational Sciences.
J.S.F. and S.E. were supported by funding from NIH/NIAID (1U19AI109713) for the “Center to develop therapeutic countermeasures to high-threat bacterial agents,” from the NIH: Centers of Excellence for Translational Research (CETR).
J.S.F. acknowledges funding from NIH/NIAID (2R42AI088893-02) and Rutgers University–NJMS.
T.L. was partially supported by the NIH award number DP7OD020317.
ABBREVIATIONS USED
- ABDT
AdaBoost
- ADME
absorption, distribution, metabolism, and excretion
- ANN
artificial neural networks
- AUC
area under the curve
- BNB
Bernoulli Naive Bayes
- CML
classic Machine Learning
- DT
Decision Tree
- DNN
Deep Neural Networks
- ECFP6
extended connectivity fingerprints of maximum diameter 6
- HTS
high throughput screening
- kNN
k-Nearest Neighbors
- QSAR
quantitative structure activity relationships
- Mtb
Mycobacterium tuberculosis
- NIAID
NIAID, National Institute of Allergy and Infectious Diseases
- PCA
Principal components analysis
- PPV
positive predicted value
- QSAR
quantitative structure activity relationships
- RF
Random forest
- ROC
receiver operating characteristic
- RP
Recursive partitioning
- SI
selectivity index
- SVM
support vector machines
- TB
Tuberculosis
- XV ROC AUC
cross-validated receiver operator characteristic curve’s area under the curve
Footnotes
Competing interests:
S.E., is owner T.L., K.M.Z., are employees and A.M.C is a consultant of Collaborations Pharmaceuticals Inc.
Supporting further details on the models, are available. This material is available free of charge via the Internet at http://pubs.acs.org. Structures of public molecules used in the training and test set and computational models for the training set and literature test set are available on Figshare with the following doi: https://doi.org/10.6084/m9.figshare.6108494.v1.
References
- 1.Global Tuberculosis Report. WHO; Geneva: 2016. [Google Scholar]
- 2.Jakab Z, Acosta CD, Kluge HH, Dara M. Consolidated Action Plan to Prevent and Combat Multidrug- and Extensively Drug-resistant Tuberculosis in the WHO European Region 2011–2015: Cost-effectiveness analysis. Tuberculosis (Edinb) 2015;95(Suppl 1):S212–S216. doi: 10.1016/j.tube.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 3.Ekins S, Spektor AC, Clark AM, Dole K, Bunin BA. Collaborative drug discovery for More Medicines for Tuberculosis (MM4TB) Drug Discov Today. 2017;22(3):555–565. doi: 10.1016/j.drudis.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikusova K, Ekins S. Learning from the past for TB drug discovery in the future. Drug Discov Today. 2017;22:534–545. doi: 10.1016/j.drudis.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riccardi G, Old IG, Ekins S. Raising awareness of the importance of funding for tuberculosis small-molecule research. Drug Discov Today. 2017;22(3):487–491. doi: 10.1016/j.drudis.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Prathipati P, Ma NL, Keller TH. Global Bayesian models for the prioritization of antitubercular agents. J Chem Inf Model. 2008;48(12):2362–70. doi: 10.1021/ci800143n. [DOI] [PubMed] [Google Scholar]
- 7.Ekins S. Progress in computational toxicology. J Pharmacol Toxicol Methods. 2014;69(2):115–40. doi: 10.1016/j.vascn.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X, Ekins S, Raufman JP, Polli JE. Computational models for drug inhibition of the human apical sodium-dependent bile acid transporter. Mol Pharm. 2009;6(5):1591–603. doi: 10.1021/mp900163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 10.Ekins S, Kaneko T, Lipinksi CA, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Ernst S, Yang J, Goncharoff N, Hohman M, Bunin B. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis. Mol BioSyst. 2010;6:2316–2324. doi: 10.1039/c0mb00104j. [DOI] [PubMed] [Google Scholar]
- 11.Ekins S, Freundlich JS. Validating new tuberculosis computational models with public whole cell screening aerobic activity datasets. Pharm Res. 2011;28:1859–69. doi: 10.1007/s11095-011-0413-x. [DOI] [PubMed] [Google Scholar]
- 12.Sarker M, Talcott C, Madrid P, Chopra S, Bunin BA, Lamichhane G, Freundlich JS, Ekins S. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium tuberculosis. Pharm Res. 2012;29:2115–2127. doi: 10.1007/s11095-012-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekins S, Reynolds R, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chem Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JSA, Bunin B. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PLOSONE. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekins S, Casey AC, Roberts D, Parish T, Bunin BA. Bayesian models for screening and TB Mobile for target inference with Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94(2):162–9. doi: 10.1016/j.tube.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekins S, Freundlich JS, Hobrath JV, Lucile White E, Reynolds RC. Combining computational methods for hit to lead optimization in Mycobacterium tuberculosis drug discovery. Pharm Res. 2014;31(2):414–35. doi: 10.1007/s11095-013-1172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekins S, Freundlich JS, Reynolds RC. Fusing dual-event datasets for Mycobacterium Tuberculosis machine learning models and their evaluation. J Chem Inf Model. 2013;53:3054–63. doi: 10.1021/ci400480s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekins S, Freundlich JS, Reynolds RC. Are Bigger Data Sets Better for Machine Learning? Fusing Single-Point and Dual-Event Dose Response Data for Mycobacterium tuberculosis. J Chem Inf Model. 2014;54:2157–65. doi: 10.1021/ci500264r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Kaushik A, Lloyd EP, Li SG, Mattoo R, Ammerman NC, Bell DT, Perryman AL, Zandi TA, Ekins S, Ginell SL, Townsend CA, Freundlich JS, Lamichhane G. Non-classical transpeptidases yield insight into new antibacterials. Nat Chem Biol. 2017;13(1):54–61. doi: 10.1038/nchembio.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekins S, Clark AM, Sarker M. TB Mobile: A Mobile App for Anti-tuberculosis Molecules with Known Targets. J Cheminform. 2013;5:13. doi: 10.1186/1758-2946-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark AM, Sarker M, Ekins S. New target predictions and visualization tools incorporating open source molecular fingerprints for TB Mobile 2.0. J Cheminform. 2014;6:38. doi: 10.1186/s13321-014-0038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djaout K, Singh V, Boum Y, Katawera V, Becker HF, Bush NG, Hearnshaw SJ, Pritchard JE, Bourbon P, Madrid PB, Maxwell A, Mizrahi V, Myllyjallio H, Ekins S. Predictive modeling targets thymidylate synthase ThyX in Mycobacterium tuberculosis. Sci Rep. 2016;6:27792. doi: 10.1038/srep27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekins S, Godbole AA, Keri G, Orfi L, Pato J, Bhat RS, Verma R, Bradley EK, Nagaraja V. Machine learning and docking models for Mycobacterium tuberculosis topoisomerase I. Tuberculosis (Edinb) 2017;103:52–60. doi: 10.1016/j.tube.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Ekins S, Pottorf R, Reynolds RC, Williams AJ, Clark AM, Freundlich JS. Looking back to the future: predicting in vivo efficacy of small molecules versus Mycobacterium tuberculosis. J Chem Inf Model. 2014;54(4):1070–82. doi: 10.1021/ci500077v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekins S, Perryman AL, Clark AM, Reynolds RC, Freundlich JS. Machine Learning Model Analysis and Data Visualization with Small Molecules Tested in a Mouse Model of Mycobacterium tuberculosis Infection (2014–2015) J Chem Inf Model. 2016;56:1332–1343. doi: 10.1021/acs.jcim.6b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark AM, Dole K, Coulon-Spector A, McNutt A, Grass G, Freundlich JS, Reynolds RC, Ekins S. Open source bayesian models: 1. Application to ADME/Tox and drug discovery datasets. J Chem Inf Model. 2015;55:1231–1245. doi: 10.1021/acs.jcim.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark AM, Ekins S. Open Source Bayesian Models: 2. Mining A “big dataset” to create and validate models with ChEMBL. J Chem Inf Model. 2015;55:1246–1260. doi: 10.1021/acs.jcim.5b00144. [DOI] [PubMed] [Google Scholar]
- 28.Clark AM, Dole K, Ekins S. Open Source Bayesian Models: 3. Composite Models for prediction of binned responses. J Chem Inf Model. 2016;56:275–285. doi: 10.1021/acs.jcim.5b00555. (J Chem Inf Model) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekins S. The next era: Deep learning in pharmaceutical research. Pharm Res. 2016;33:2594–603. doi: 10.1007/s11095-016-2029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korotcov A, Tkachenko V, Russo DP, Ekins S. Comparison of Deep Learning With Multiple Machine Learning Methods and Metrics Using Diverse Drug Discovery Data Sets. Mol Pharm. 2017;14(12):4462–4475. doi: 10.1021/acs.molpharmaceut.7b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100-7. doi: 10.1093/nar/gkr777. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–90. doi: 10.1093/nar/gkt1031. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anon ChEMBL URL. https://www.ebi.ac.uk/chembl/faq#faq35.
- 34.http://molmatinf.com/fmtdatasheet.html
- 35.Clark A. XMDS. https://cheminf20.org/category/xmds/
- 36.Anon Assay Central video. https://www.youtube.com/watch?v=aTJJ6Tyu4bY&feature=youtu.be.
- 37.Ananthan S, Faaleolea ER, Goldman RC, Hobrath JV, Kwong CD, Laughon BE, Maddry JA, Mehta A, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Shindo N, Showe DN, Sosa MI, Suling WJ, White EL. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddry JA, Ananthan S, Goldman RC, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Sosa MI, White EL, Zhang W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds RC, Ananthan S, Faaleolea E, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Sosa MI, Thammasuvimol E, White EL, Zhang W, Secrist JA., 3rd High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2(6):662–7. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 41.Heifets LB, Flory MA, Lindholm-Levy PJ. Does pyrazinoic acid as an active moiety of pyrazinamide have specific activity against Mycobacterium tuberculosis? Antimicrob Agents Chemother. 1989;33(8):1252–4. doi: 10.1128/aac.33.8.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimhony O, Cox JS, Welch JT, Vilcheze C, Jacobs WR., Jr Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat Med. 2000;6(9):1043–7. doi: 10.1038/79558. [DOI] [PubMed] [Google Scholar]
- 43.Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, 3rd, Wang H, Zhang W, Zhang Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333(6049):1630–2. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52(5):790–5. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 45.Shi W, Chen J, Feng J, Cui P, Zhang S, Weng X, Zhang W, Zhang Y. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg Microbes Infect. 2014;3(8):e58. doi: 10.1038/emi.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon NA, Peterson ND, Rosen BC, Baughn AD. Pantothenate and pantetheine antagonize the antitubercular activity of pyrazinamide. Antimicrob Agents Chemother. 2014;58(12):7258–63. doi: 10.1128/AAC.04028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gopal P, Yee M, Sarathy J, Low JL, Sarathy JP, Kaya F, Dartois V, Gengenbacher M, Dick T. Pyrazinamide Resistance Is Caused by Two Distinct Mechanisms: Prevention of Coenzyme A Depletion and Loss of Virulence Factor Synthesis. ACS Infect Dis. 2016;2(9):616–626. doi: 10.1021/acsinfecdis.6b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gopal P, Tasneen R, Yee M, Lanoix JP, Sarathy J, Rasic G, Li L, Dartois V, Nuermberger E, Dick T. In Vivo-Selected Pyrazinoic Acid-Resistant Mycobacterium tuberculosis Strains Harbor Missense Mutations in the Aspartate Decarboxylase PanD and the Unfoldase ClpC1. ACS Infect Dis. 2017;3(7):492–501. doi: 10.1021/acsinfecdis.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karapetyan K, Batchelor C, Sharpe D, Tkachenko V, Williams AJ. The Chemical Validation and Standardization Platform (CVSP): large-scale automated validation of chemical structure datasets. J Cheminform. 2015;7:30. doi: 10.1186/s13321-015-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carletta J. Assessing agreement on classification tasks: The kappa statistic. Computational Linguistics. 1996;22:249–254. [Google Scholar]
- 51.Cohen J. A coefficient of agreement for nominal scales. Education and Psychological Measurement. 1960;20:37–46. [Google Scholar]
- 52.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405(2):442–51. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 53.Zhao H, Lu Y, Sheng L, Yuan Z, Wang B, Wang W, Li Y, Ma C, Wang X, Zhang D, Huang H. Discovery of Fluorine-Containing Benzoxazinyl-oxazolidinones for the Treatment of Multidrug Resistant Tuberculosis. ACS Med Chem Lett. 2017;8(5):533–537. doi: 10.1021/acsmedchemlett.7b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tseng CH, Tung CW, Wu CH, Tzeng CC, Chen YH, Hwang TL, Chen YL. Discovery of Indeno[1,2-c]quinoline Derivatives as Potent Dual Antituberculosis and Anti-Inflammatory Agents. Molecules. 2017;22(6) doi: 10.3390/molecules22061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surase YB, Samby K, Amale SR, Sood R, Purnapatre KP, Pareek PK, Das B, Nanda K, Kumar S, Verma AK. Identification and synthesis of novel inhibitors of mycobacterium ATP synthase. Bioorg Med Chem Lett. 2017;27(15):3454–3459. doi: 10.1016/j.bmcl.2017.05.081. [DOI] [PubMed] [Google Scholar]
- 56.Rodrigues Felix C, Gupta R, Geden S, Roberts J, Winder P, Pomponi SA, Diaz MC, Reed JK, Wright AE, Rohde KH. Selective Killing of Dormant Mycobacterium tuberculosis by Marine Natural Products. Antimicrob Agents Chemother. 2017;61(8) doi: 10.1128/AAC.00743-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ojima I, Awasthi D, Wei L, Haranahalli K. Strategic incorporation of fluorine in the drug discovery of new-generation antitubercular agents targeting bacterial cell division protein FtsZ. J Fluor Chem. 2017;196:44–56. doi: 10.1016/j.jfluchem.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lv K, You X, Wang B, Wei Z, Chai Y, Wang B, Wang A, Huang G, Liu M, Lu Y. Identification of Better Pharmacokinetic Benzothiazinone Derivatives as New Antitubercular Agents. ACS Med Chem Lett. 2017;8(6):636–641. doi: 10.1021/acsmedchemlett.7b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu X, Tang J, Liu Z, Li M, Zhang T, Zhang X, Ding K. Discovery of new chemical entities as potential leads against Mycobacterium tuberculosis. Bioorg Med Chem Lett. 2016;26(24):5916–5919. doi: 10.1016/j.bmcl.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Hong WD, Gibbons PD, Leung SC, Amewu R, Stocks PA, Stachulski A, Horta P, Cristiano MLS, Shone AE, Moss D, Ardrey A, Sharma R, Warman AJ, Bedingfield PTP, Fisher NE, Aljayyoussi G, Mead S, Caws M, Berry NG, Ward SA, Biagini GA, O’Neill PM, Nixon GL. Rational Design, Synthesis, and Biological Evaluation of Heterocyclic Quinolones Targeting the Respiratory Chain of Mycobacterium tuberculosis. J Med Chem. 2017;60(9):3703–3726. doi: 10.1021/acs.jmedchem.6b01718. [DOI] [PubMed] [Google Scholar]
- 61.He C, Preiss L, Wang B, Fu L, Wen H, Zhang X, Cui H, Meier T, Yin D. Structural Simplification of Bedaquiline: the Discovery of 3-(4-(N,N-Dimethylaminomethyl)phenyl)quinoline-Derived Antitubercular Lead Compounds. ChemMedChem. 2017;12(2):106–119. doi: 10.1002/cmdc.201600441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes MN, Braga RC, Grzelak EM, Neves BJ, Muratov E, Ma R, Klein LL, Cho S, Oliveira GR, Franzblau SG, Andrade CH. QSAR-driven design, synthesis and discovery of potent chalcone derivatives with antitubercular activity. Eur J Med Chem. 2017;137:126–138. doi: 10.1016/j.ejmech.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia X, Maliski EG, Gallant P, Rogers D. Classification of kinase inhibitors using a Bayesian model. J Med Chem. 2004;47(18):4463–70. doi: 10.1021/jm0303195. [DOI] [PubMed] [Google Scholar]
- 64.Zadrozny B, Elkan C. Obtaining calibrated probability estimates from decision trees and naive Bayesian classifiers; ICML ‘01 proceedings of the Eighteenth International Conference on Machine Learning; June 28–July 1; 2001. pp. 609–616. [Google Scholar]
- 65.Niculescu-Mizil A, Caruana R. Predicting good probabilities with supervised learning; ICML ‘05 Proceedings of the 22nd International conference on Machine Learning, Bonn, Germany; Bonn, Germany. 2005. pp. 625–632. [Google Scholar]
- 66.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2(3) [Google Scholar]
- 67.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Capuzzi SJ, Politi R, Isayev O, Farag S, Tropsha A. QSAR Modeling of Tox21 Challenge Stress Response and Nuclear Receptor Signaling Toxicity Assays. Frontiers in Environmental Science. 2016;4(3) [Google Scholar]
- 69.Russakovsky O, deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. ImageNet large scale visual recognition challenge. Int J Comp Vision. 2015;115(3):211–252. [Google Scholar]
- 70.Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A. Deep Learning Applications for Predicting Pharmacological Properties of Drugs and Drug Repurposing Using Transcriptomic Data. Mol Pharm. 2016;13(7):2524–30. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of Deep Learning in Biomedicine. Mol Pharm. 2016;13(5):1445–54. doi: 10.1021/acs.molpharmaceut.5b00982. [DOI] [PubMed] [Google Scholar]
- 72.Jing Y, Bian Y, Hu Z, Wang L, Xie XS. Deep Learning for Drug Design: an Artificial Intelligence Paradigm for Drug Discovery in the Big Data Era. AAPS J. 2018;20(3):58. doi: 10.1208/s12248-018-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018 doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, Tan J, Han D, Zhu H. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today. 2017;22(11):1680–1685. doi: 10.1016/j.drudis.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 75.Gawehn E, Hiss JA, Schneider G. Deep Learning in Drug Discovery. Mol Inform. 2016;35(1):3–14. doi: 10.1002/minf.201501008. [DOI] [PubMed] [Google Scholar]
- 76.Gamo F-J, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera J-L, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 77.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jimenez-Diaz MB, Martinez MS, Wilson EB, Tripathi AK, Gut J, Sharlow ER, Bathurst I, El Mazouni F, Fowble JW, Forquer I, McGinley PL, Castro S, Angulo-Barturen I, Ferrer S, Rosenthal PJ, Derisi JL, Sullivan DJ, Lazo JS, Roos DS, Riscoe MK, Phillips MA, Rathod PK, Van Voorhis WC, Avery VM, Guy RK. Chemical genetics of Plasmodium falciparum. Nature. 2010;465(7296):311–5. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inoyama D, Paget SD, Russo R, Kandasamy S, Kumar P, Singleton E, Occi J, Tuckman M, Zimmerman MD, Ho HP, Perryman AL, Dartois V, Connell N, Freundlich JS. Novel Pyrimidines as Antitubercular Agents. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.02063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perryman AL, Yu W, Wang X, Ekins S, Forli S, Li SG, Freundlich JS, Tonge PJ, Olson AJ. A Virtual Screen Discovers Novel, Fragment-Sized Inhibitors of Mycobacterium tuberculosis InhA. J Chem Inf Model. 2015 doi: 10.1021/ci500672v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2002;46(8):2720–2. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anon CDD Public. https://www.collaborativedrug.com/pages/public_access.
- 82.Soro O, Pesce A, Raggi M, Debbia EA, Schito GC. Selection of rifampicin-resistant Mycobacterium tuberculosis does not occur in the presence of low concentrations of rifaximin. Clin Microbiol Infect. 1997;3(1):147–151. doi: 10.1111/j.1469-0691.1997.tb00272.x. [DOI] [PubMed] [Google Scholar]
- 83.Zang Q, Mansouri K, Williams AJ, Judson RS, Allen DG, Casey WM, Kleinstreuer NC. In Silico Prediction of Physicochemical Properties of Environmental Chemicals Using Molecular Fingerprints and Machine Learning. J Chem Inf Model. 2017;57(1):36–49. doi: 10.1021/acs.jcim.6b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Z, Ramsundar B, Feinberg EN, Gomes J, Geniesse C, Pappu AS, Leswing K, Pande V. MoleculeNet: A Benchmark for Molecular Machine Learning. doi: 10.1039/c7sc02664a. https://arxiv.org/ftp/arxiv/papers/1703/1703.00564.pdf. [DOI] [PMC free article] [PubMed]
- 85.Vock DM, Wolfson J, Bandyopadhyay S, Adomavicius G, Johnson PE, Vazquez-Benitez G, O’Connor PJ. Adapting machine learning techniques to censored time-to-event health record data: A general-purpose approach using inverse probability of censoring weighting. J Biomed Inform. 2016;61:119–31. doi: 10.1016/j.jbi.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015;16(6):321–32. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mitchell JB. Machine learning methods in chemoinformatics. Wiley Interdiscip Rev Comput Mol Sci. 2014;4(5):468–481. doi: 10.1002/wcms.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stalring JC, Carlsson LA, Almeida P, Boyer S. AZOrange - High performance open source machine learning for QSAR modeling in a graphical programming environment. J Cheminform. 2011;3:28. doi: 10.1186/1758-2946-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheridan RP. Time-split cross-validation as a method for estimating the goodness of prospective prediction. J Chem Inf Model. 2013;53(4):783–90. doi: 10.1021/ci400084k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.