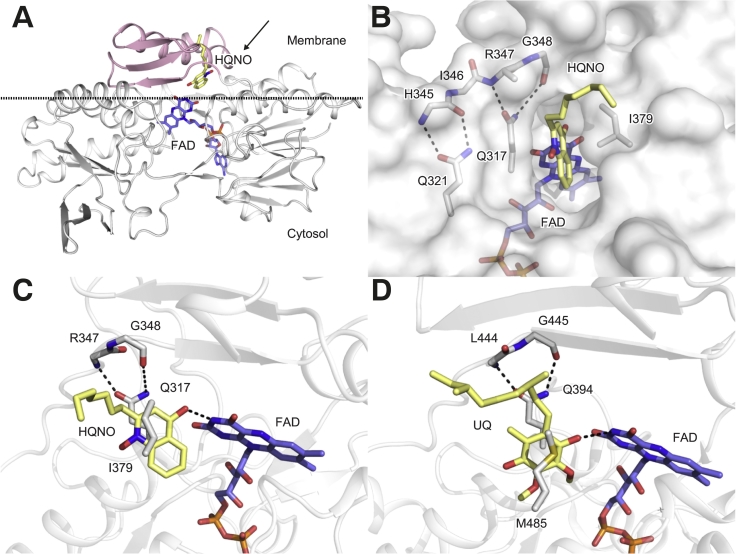
Fig. 4.
Crystal structure of the NDH-2–HQNO complex. A) An overview of the NDH-2–HQNO complex structure showing the location of the membrane interface (dashed-line), FAD (blue) and HQNO (yellow). The C-terminal membrane anchoring domain is coloured in pink. B) A top view of a panel A (arrowed) showing the quinone-binding site from the membrane side. A pair of Q317 and I379 side chains (white) clamp a quinone-head group of HQNO (yellow). Two conserved glutamate residues (Q317 and Q321) in the AQXAXQ motif holding the linker that separates the NADH- and quinone-binding sites (hydrogen bonds are shown with dashed lines). C) A side view of panel B showing a clamped HQNO head group and a hydrogen bond between the O1 atom of the HQNO quinone head group and the N3 atom of FAD with a distance of 2.8 Å. D) The binding of the quinone head group is conserved for the yeast Ndi1. In the yeast Ndi1–ubiquinone (UQ1) complex structure (PDB: 4G73), a UQ (yellow) head group is clamped with Q394 and M485 and also forms a hydrogen bond with FAD (blue).