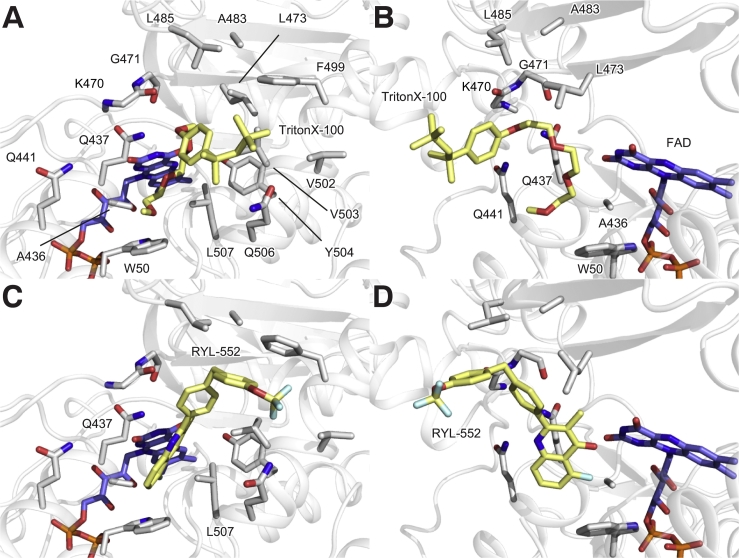
Fig. 7.
In silico docking models of the PfNDH-2–Triton X100 complex and PfNDH-2–RYL552 complex. A) In silico docking model of the PfNDH-2–Triton X100 complex. FAD and Triton X-100 molecules are represented as blue and yellow stick models. Residues involved in the hydrophobic groove of the C-terminal membrane anchoring domain are also represented as stick models. B) A view of panel A rotated by 90° clockwise. The first amphipathic helix is omitted for clarity. C) In silico docking model of the PfNDH-2–RYL552 complex. FAD and RYL-552 molecules are represented as blue and yellow stick models. Residues involved in the hydrophobic groove of the C-terminal membrane anchoring domain are also represented as stick models. D) A view of panel C rotated by 90° clockwise. The first amphipathic helix is omitted for clarity. Residue labels are omitted for panels C and D as the images are identical to those in panels A and B.