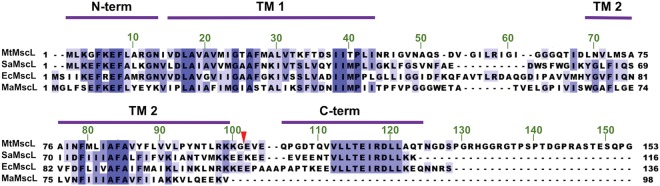
Figure 1.
Alignment of MscL sequences. Full-length sequences of Mycobacterium tuberculosis MscL, Staphylococcus aureus MscL, Escherichia coli MscL and Methanosarcina acetivorans MscL were aligned using Clustal Omega. The sequence identities between pairs of sequences are MtMscL to SaMscL 40.5%, MtMscL to EcMscL 35.2%, and MtMscL to MaMscL 29.2%. The red triangle over the sequence alignment depicts the location of the (C) terminal truncation after residue 101 of MtMscLΔC. The blue shadings represent sequence identity gradients. The locations of the secondary structure regions are labeled based on the structure for MtMscL, PDBID 2OAR. The residue number legend in green along the top of the alignment corresponds to MtMscL, while the residue numbers on either side of the sequences correspond to that particular homolog. High variability is evident in the periplasmic loop among the homologs.