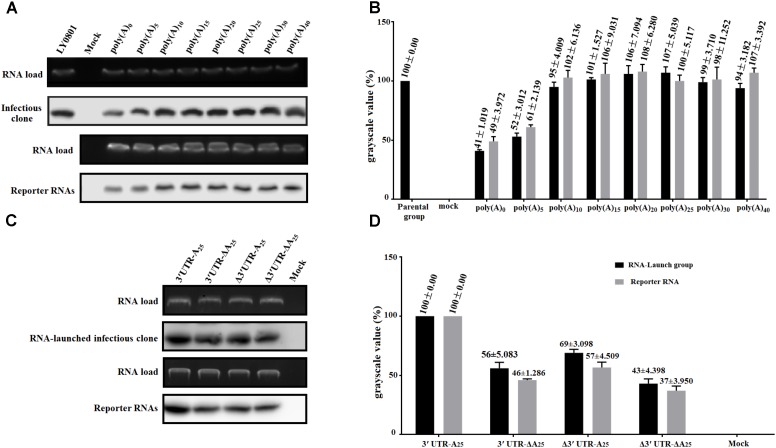
FIGURE 2.
Effects of poly(A) tail length and 3′ UTR on RNA stability. DEFs transfected with 0.8 μg of the in vitro transcribed RNA were collected at 4 hpt and were immediately used for total RNA extraction. The samples were analyzed by 1.5% formaldehyde-agarose gel electrophoresis and autoradiography. Digoxigenin (DIG)-labeled RNA probes (based on 81–351 of LY0801) was used to in this assay. The gels show the amount of input RNA remaining at 4 hpt. (A) The remaining amount of pR-DHAV-1-An or pDHAV-1-An was measured at 4 hpt. (B) The grayscale value of represent the mean ± standard deviation of three replicate experiments for poly(A) n group. The grayscale value of parental group was set as 100%. (C) The remaining amount of complete/mutated RNA-launched infectious clone or reporter RNA was measured at 4 hpt. The upper panel shows the ethidium bromide staining of the agarose gel loaded with aliquots of 800 ng RNAs. (D) The average grayscale values of three replicate experiments for each group. The grayscale value of pR-DHAV-1 or pDHAV-3′UTR-A25 was set as 100%.