Figure 1. Overexpression of S.aureus gpsB inhibits cell division in B. subtilis.
(A) Luria-Bertani agar plates streaked with wild type B. subtilis (WT, strain PY79), or otherwise wild type B. subtilis harboring an inducible copy of gpsBBs (GG18), gpsBBs-gfp (GG19), gpsBSa (GG7), or gpsBSa-gfp (GG8) integrated into the chromosome, in the absence (left) or presence (right) of inducer. (B–M) Morphology of cells of different deletion mutants of B. subtilis (ΔezrA, GG35; ΔponA, CS26; ΔprkC, CS24; ΔgpsB, CS40; ΔdivIVA, CS94) harboring an inducible copy of gpsBSa grown in the absence (B, D, F; H, J, L) or presence (C, E, G, I, K, M) of inducer. (N–O) Localization of FtsZ-GFP in a strain (GG9) harboring an inducible copy of gpsBSa grown in the absence (N) or presence (O) of inducer. Membranes (red; B–O) visualized using the fluorescent dye FM4-64; chromosomes (blue; B–M) visualized using DAPI; FtsZ-GFP localization (green; N–O). Scale bar: 1 μm. Genotypes are listed in Key Resources Table.
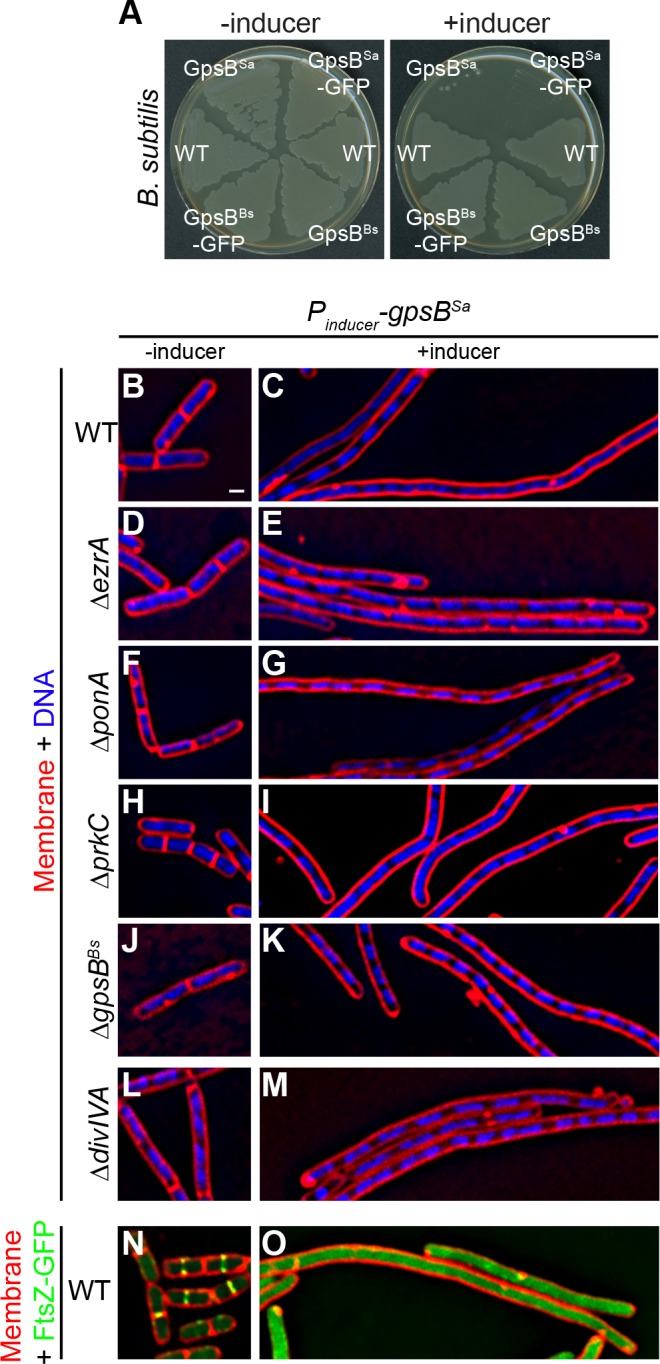
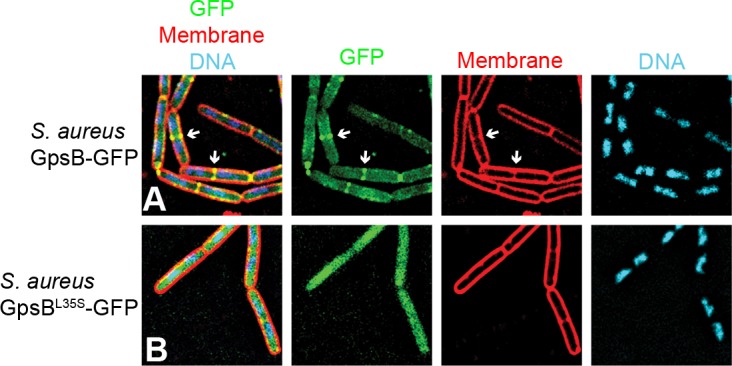
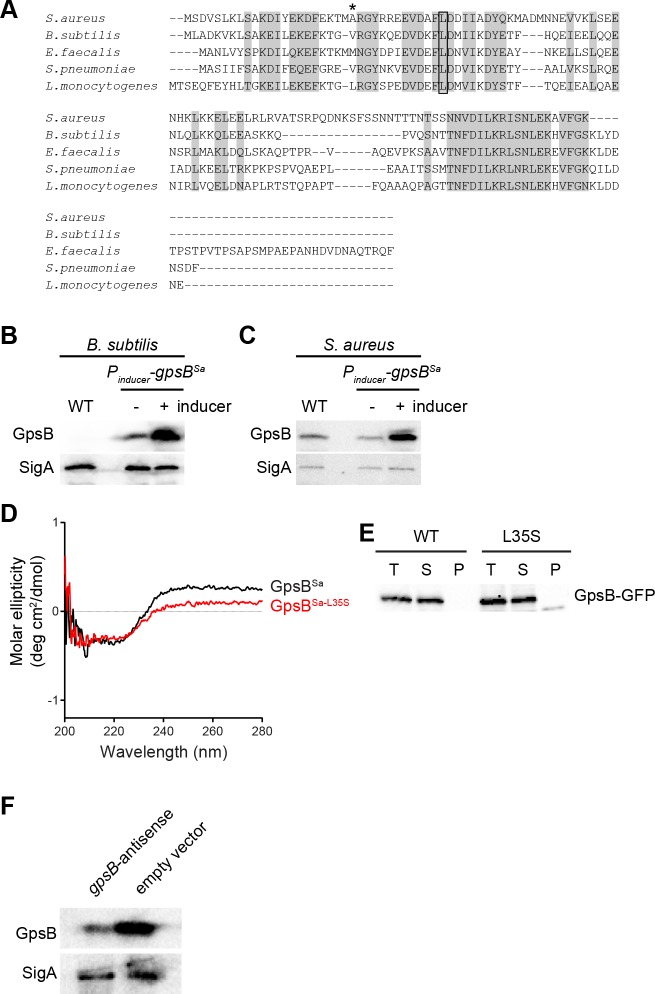
Figure 1—figure supplement 1. GpsB sequence and subcellular distribution in S.aureus.
Figure 1—figure supplement 2. GpsBSa-GFP localizes to division septa in B.subtilis.