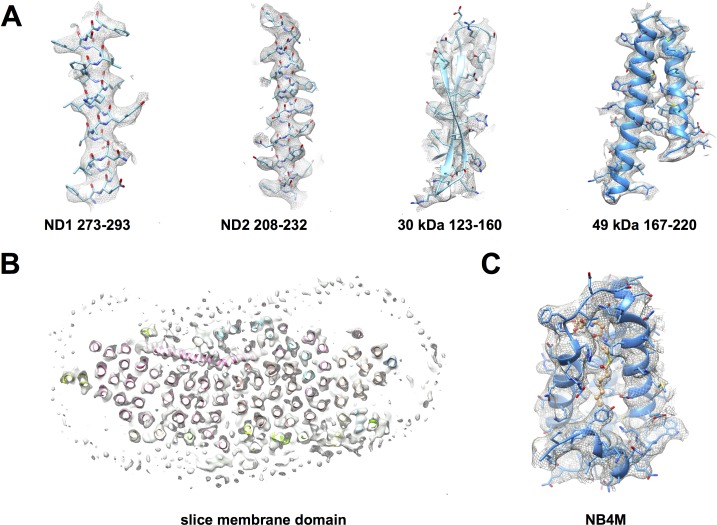
Figure 3. Cryo-EM map of deactive complex I with fitted models.
(A) Selected regions of the matrix domain. (B) A horizontal cross-section through the membrane arm shows TMH fits. (C) A region of the accessory LYR protein subunit NB4M (Angerer et al., 2017); the acyl chain appended to the phosphopantetheine group of the adjacent acyl carrier protein ACPM1 inserts into the interior of NB4M. Cofactor and acyl chain are drawn as ball-and-stick model.