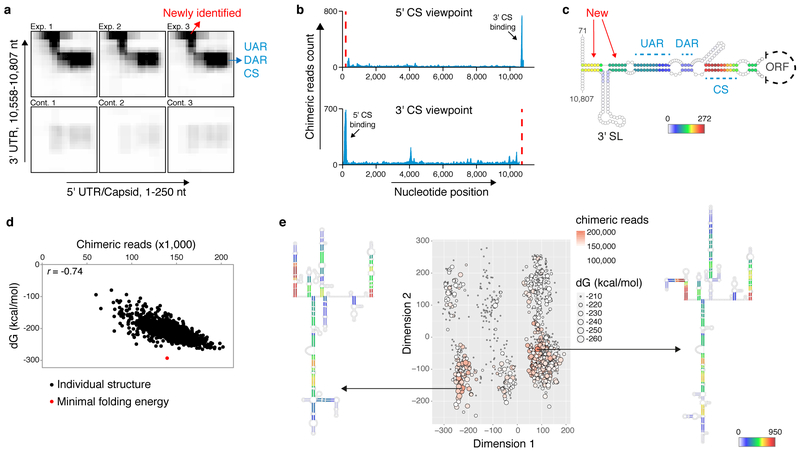
Fig. 2: The ZIKV genomic structure inside human cells.
a, Heatmap of RNA-RNA interactions between cyclization elements. Exp: experiment; cont: control. b, Viewpoint histograms showing binding positions of the cyclization sequences along the ZIKV genome. Viewpoint regions are marked by dashed red lines. c, Probed interactions along the circular genome conformation. Colour code indicates the number of non-redundant chimeric reads supporting each base-pair. New: newly identified base-pairing. d, Folding energy (dG) and experimentally supporting evidence (chimera reads) for each of the 1,000 computationally predicted structures corresponding to ZIKV genome nt 2,288 - 3,323. r: Pearson correlation coefficient. e, Clustering and prediction of alternative conformations for the region shown in (d). Colour code as described in (c).