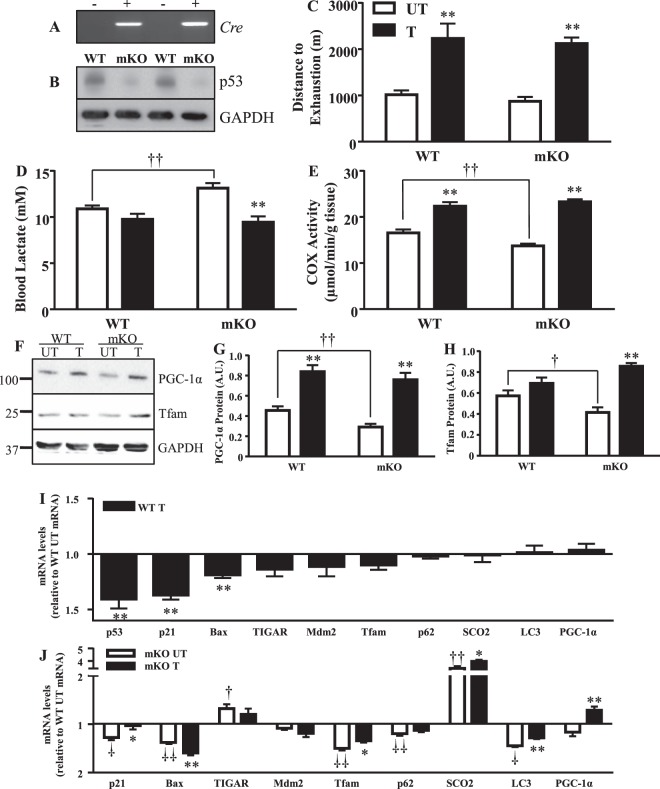
Figure 2.
Mitochondrial content, exercise capacity, and lactate handling improves with exercise training, while gene expression is altered. The deletion of p53 in skeletal muscle of MS mKO mice was examined through (A) genotype against the Cre transcript, and (B) total p53 protein in whole muscle. Following the 6-week training/sedentary protocol, mice were subjected to an exhaustive bout of exercise to determine training-induced adaptations by measuring (C) distance to exhaustion (n = 6–8/group); **p ≤ 0.01, UT vs. T, 2-way ANOVA, and (D) final lactate production levels (n = 6–8/group), **p ≤ 0.01, UT vs. T; ††p ≤ 0.01, UT WT vs. mKO, Student’s t-test. Mitochondrial biogenesis with training was measured through the assessment of (E) COX enzyme activity, a marker of mitochondrial content (n = 6–8/group); **p ≤ 0.01, UT vs. T, 2-way ANOVA; ††p ≤ 0.01, UT WT vs. mKO, Student’s t-test, and (F) mitochondrial biogenesis markers (G) PGC-1α protein (n = 5–8/group); **p ≤ 0.01, UT vs. T, 2-way ANOVA; ††p ≤ 0.01, UT WT vs. mKO, Student’s t-test, and (H) Tfam protein (n = 6–7/group); **p ≤ 0.01, UT vs. T, 2-way ANOVA; †p ≤ 0.05, UT WT vs. mKO, Student’s t-test. (I) The effect of training on mRNA transcripts of signaling pathways including cellular senescence (p21), apoptosis (Bax), metabolism (TIGAR), autophagy (p62, LC3), oxidative phosphorylation (SCO2), mitochondrial biogenesis (PGC-1α, Tfam) and p53 and its negative regulator Mdm2, in WT mice. Data are presented as fold change over WT control levels (n = 8–10/group); **p ≤ 0.01, UT vs. T, Student’s t-test. (J) Effect of the absence of p53, and training on mRNA transcripts of signaling pathways. Data presented as fold change of mKO over WT untrained levels and as mKO trained over untrained levels (n = 7–11/group); *p ≤ 0.05, **p ≤ 0.01, mKO UT vs. T; †p ≤ 0.05, ††p ≤ 0.01, UT WT vs. mKO, Student’s t-test. Data are presented as mean ± SEM.