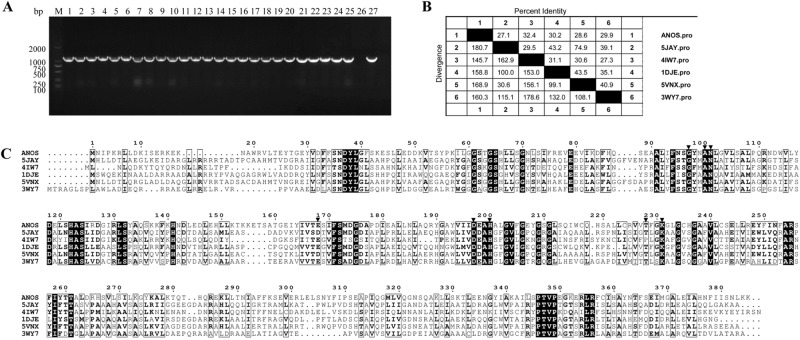
Figure 1.
Analyses of Riemerella anatipestifer AS87_RS09170 gene and deduced amino acid sequence. (A) Polymerase chain reaction (PCR) analysis. The AS87_RS09170 gene was amplified from all R. anatipestifer strains tested. Lane M, DL2000 DNA Marker (Takara); lanes 1–5: R. anatipestifer serotype 1 strains GTB1, GTB2, CQ1, CQ3 and NJ4; lanes 6–10, R. anatipestifer serotype 2 strains GDO-3, JY1, JY2, NJ3 and SC2; lanes 11–15, R. anatipestifer serotype 10 strains GDO-1, HXb2, YXb1, YXb11 and YXL1; lanes 16–20, R. anatipestifer serotype 15 strains SQ007, SQ003, SQ004, YGT002 and MA001; lanes 21–25, R. anatipestifer undefined serotype strains GDO-6, GDO-7, G46, G77 and JY-6; lane 26, negative control of Yb2ΔbioF; lane 27, positive control of Yb2. A 1158-bp fragment was amplified from all 25 R. anatipestifer strains tested. The full-length gel is presented in Supplementary Figure S2. (B) Identity and divergence analysis. The predicted R. anatipestifer 8-amino-7-oxononanoate synthase (AONS) and other known AONS were compared. The crystallized AONS sequences [Paraburkholderia xenovorans (pdb5JAY), Francisella tularensis (pdb4IW7), Escherichia coli (pdb1DJE), Burkholderia multivorans (pdb5VNX) and Mycobacterium smegmatis (pdb3WY7)] were retrieved from the Uniprot database and later aligned with Clustal W algorithm in the MegAlign program from the DNASTAR Lasergene suite. (C) Multiple sequence alignment. Strictly conserved residues in all sequences are boxed in black. Residues forming the conserved pyridoxal 5′-phosphate binding pocket are indicated by a black inverted triangle (G97, Y98, N101, E168, D197, H200, T228 and K231).