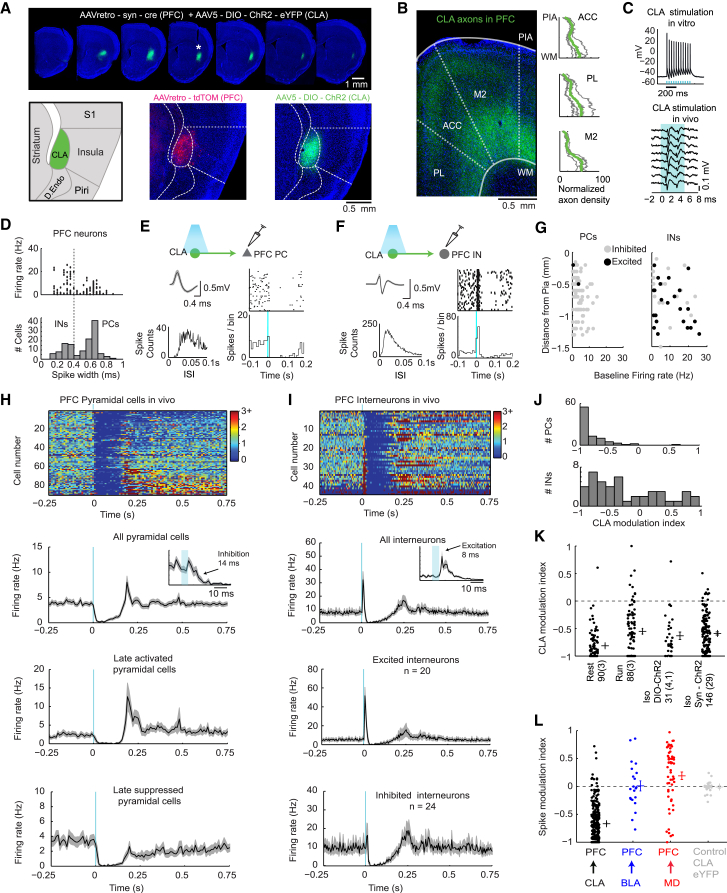
Figure 1.
Claustrocortical Projections Control the PFC through Uniquely Strong Feedforward Inhibition In Vivo
(A) Top, the distribution of CLA ChR2 virus labeling in the rostro-caudal axis, following injection of AAVretro-syn-Cre into the PFC together with AAV5-DIO-ChR2-eYFP into the CLA. The white asterisk shows the location of the optical fiber. Bottom, a schematic showing the brain regions neighboring the CLA, the retrograde labeling of CLA neurons with tdTomato following injection of AAVretro-CAG-tdTomato in the PFC (red) and ChR2 labeling of claustrocortical neurons (green).
(B) A representative image showing CLA axons in the PFC, and axon density measurements taken in the PFC regions of ACC, PL, and M2. Individual animals (gray) and group mean (green, n = 4) are shown.
(C) Example CLA neurons responding with spiking to 473 nm 5 ms light pulses delivered to the CLA in vitro (above) or in vivo (below).
(D) A scatterplot of the firing rate versus spike width, and the histogram of spike waveform widths for all (PCs) and interneurons (INs).
(E) An example spike waveform, interspike interval histogram (ISI), and peri-stimulus time histogram (PSTH) for an example PC in response to optogenetic CLA activation (blue line).
(F) The same as in (E), but for an example PFC interneuron.
(G) The firing rate distribution of PCs and INs as a function of depth measured from the pia. Neurons excited by CLA activation are indicated in black, and inhibited cells are indicated in gray.
(H) Top, the mean normalized firing rate for all PCs in response to optogenetic stimulation of the CLA in awake mice (top). Below, the mean firing rate for all cells, and the subset of cells showing late inhibition or activation following CLA activation. The inset in the mean PSTH shows a magnified view of the group mean PSTH.
(I) Top, the mean normalized firing rate for all INs in response to optogenetic stimulation of the CLA in awake mice (top). Below, the mean firing rate for all INs, and INs which were excited or inhibited. The inset in the mean PSTH shows a magnified view of the group mean PSTH.
(J) Histogram of the single cell modulation index (MI, see STAR Methods) for PCs and INs following the CLA pulse when the animal was at rest.
(K) The comparison of the CLA modulation index for PCs across behavioral states and virus types. Cells are shown during rest, during locomotion (Run), and during isoflurane anesthesia (Iso, with AAV5-DIO-ChR2 or AAV1-syn-ChR2). The values indicate number of cells (and number of animals). In the case of DIO-ChR2, four mice and one rat were used.
(L) The comparison between the strength and direction of PFC modulation by the CLA (n = 37 mice, 267 single-unit and multi-unit recordings), BLA (n = 2 mice, 21 multi-unit recordings), and MD (n = 4 rats, 31 single and multi-unit recordings, and n = 2 mice, 26 single and multi-unit recordings). The data from mice and rats for MD experiments were pooled, as the mean MI for rats (0.21 ± 0.08) was not significantly different than for mice (0.15 ± 0.12, p = 0.6). Control experiments with CLA–eYFP are also shown (n = 2 mice, 24 recordings). Interneuron recordings were excluded from all analyses here except the control recordings. Data are expressed as mean ± SEM. See also Figure S1.