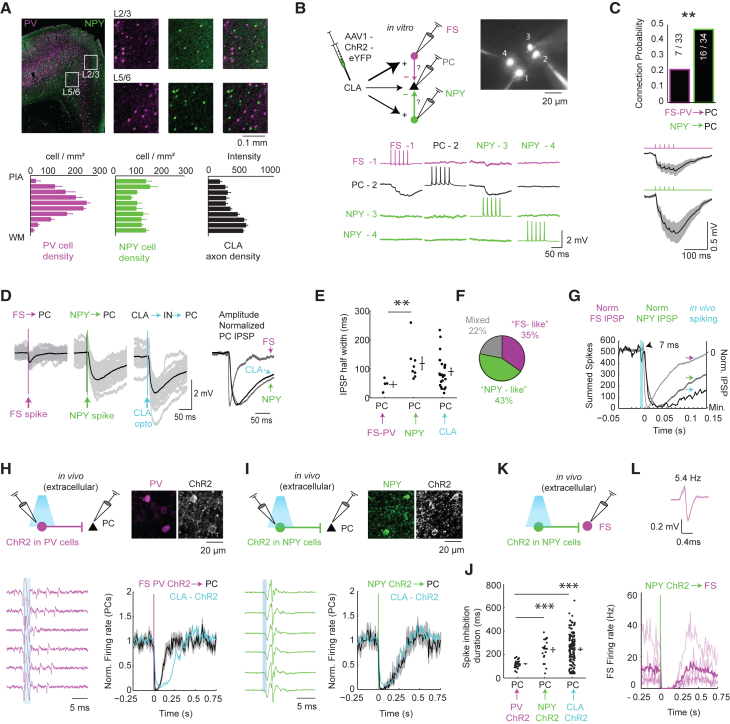
Figure 3.
The Comparison between NPY- and PV-Mediated Inhibition in the PFC
(A) Anatomical characterization of the layer dependence of PV and NPY neurons in the PFC. An example image is shown. Areas denoted by the white boxes are expanded on the right. The histograms below show the mean ± SEM of the cell density as a function of the normalized distance from the pia to white matter (WM). The density of CLA axons as a function of distance from pia is also shown. Densities were averaged across the prelimbic cortex, anterior cingulate cortex, and secondary motor cortex (n = 4 mice). PV cells were identified immunohistochemically (magenta) in NPY-GFP mice.
(B) A schematic showing the comparison between feedforward inhibition arising from either NPY or FS cells in vitro. Patch-clamp recordings were made from FS-PV cells, NPY cells, and PCs. An example quadruple-patch experiment where a FS, PC, and two NPY cells were simultaneously recorded, and the connectivity between cells, is shown during 50 Hz pulse trains.
(C) The connection probability for pairs of FS cells and PCs and for NPY cells and PCs. The mean IPSP in response to FS or NPY stimulation is shown below.
(D) An example experiment showing the unitary monosynaptic IPSP from an FS cell to PC, from a NPY cell to PC, and the IPSP in response to optogenetic CLA stimulation. Gray traces throughout are individual trials. For unitary connections, one spike was elicited in the presynaptic cell. The amplitude-normalized responses are shown on the right. Note the similarity between the NPY→PC IPSP and the CLA-mediated IPSP in this particular experiment.
(E) The decay time half-widths for IPSPs elicited by FS cells, by NPY cells, and by CLA stimulation. The CLA inhibitory kinetics span a range of values in vitro. However, the CLA-mediated IPSP half-width was more similar to the NPY than FS values, and NPY-mediated IPSPs were slower than FS-mediated IPSPs.
(F) The pie chart shows the proportion of CLA-evoked IPSPs that were classified as “NPY-like” or “FS-like,” or mixed based on IPSP rise times and IPSP half-widths.
(G) FS and NPY IPSPs overlaid on the in vivo spiking response to CLA stimulation. IPSPs were shifted to 7 ms to match the predicted onset of IPSPs in vivo based on the observation that interneurons began spiking ∼7 ms following CLA stimulation.
(H) In vivo optogenetic activation of PV interneurons suppresses PC neurons. An example putative opto-tagged PV neuron in the PFC responding to 2 ms, 473 nm light pulses. The cell fired in one to three spike bursts similar to many interneurons recorded during CLA activation (Figure S1). On the right is the mean response of PCs to PV activation (black) and the mean response in all cells inhibited by CLA activation (blue). Dark lines show the mean, and shaded regions show the SEM (n = 3 mice, 34 PC recordings).
(I) In vivo optogenetic activation of NPY interneurons suppresses PC neurons. An example putative opto-tagged NPY neuron in the PFC responding to 1 ms of 473 nm light pulses. The cell fired one spike per trial. On the right is the mean response of PCs to NPY activation (black) and the mean response in all cells inhibited by CLA activation (blue). Dark lines show the mean and shaded regions the SEM (n = 2 mice, 22 PC recordings).
(J) The duration of spike inhibition in PV-ChR2, NPY-ChR2, and CLA activation experiments. Spike inhibition duration was measured by the time elapsed between falling below 50% of the baseline firing rate and recovering back to 50% of the baseline firing (n = 32 mice; 155 recordings for CLA activation).
(K) Schematic depicting the experiment in which NPY cells are activated with ChR2 while recording from putative FS interneurons.
(L) The spike shape of an extracellular recording of a putative FS cell (above) that was identified during NPY-ChR2 activation. The baseline firing rate of this neuron is indicated above the waveform. The PSTHs for all experiments are shown below (n = 5). The dark line is the mean, and light shaded lines are individual experiments. Data are expressed as mean ± SEM throughout.
∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S3.