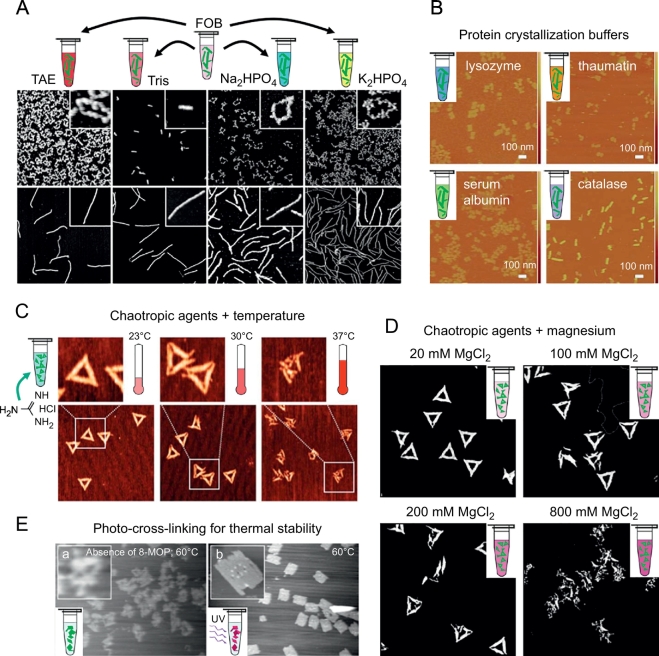
Fig. 1.
DNA origami under different biophysical and biochemical conditions. (A)24HBs (top panel) and 6HBs (bottom panel) in different low-magnesium buffers (FOB denotes folding buffer) [52]. (B)Tubular DNA origami structures in protein crystallization buffers of lysozyme, thaumatin, serum albumin and catalase [57]. (C)DNA origami triangles incubated at different temperatures in the presence of the chaotropic agent GdmCl at 6M concentration [61]. (D)DNA origami triangles in 4M GdmCl with varying MgCl2 concentrations [62]. (E)DNA origami sheets without (left) and with (right) 8-MOP-based photo-cross-linking at 60 °C [63]. (A)is reproduced with permission from Ref. [52]; copyright John Wiley & Sons 2018. (B)is reproduced with permission from Ref. [57]; published by Royal Society of Chemistry 2015. (C)is reproduced with permission from Ref. [61]; published by Royal Society of Chemistry 2016. (D)is reproduced with permission from Ref. [62]; copyright John Wiley & Sons 2017. (E)is reproduced with permission from Ref. [63]; copyright American Chemical Society 2011.