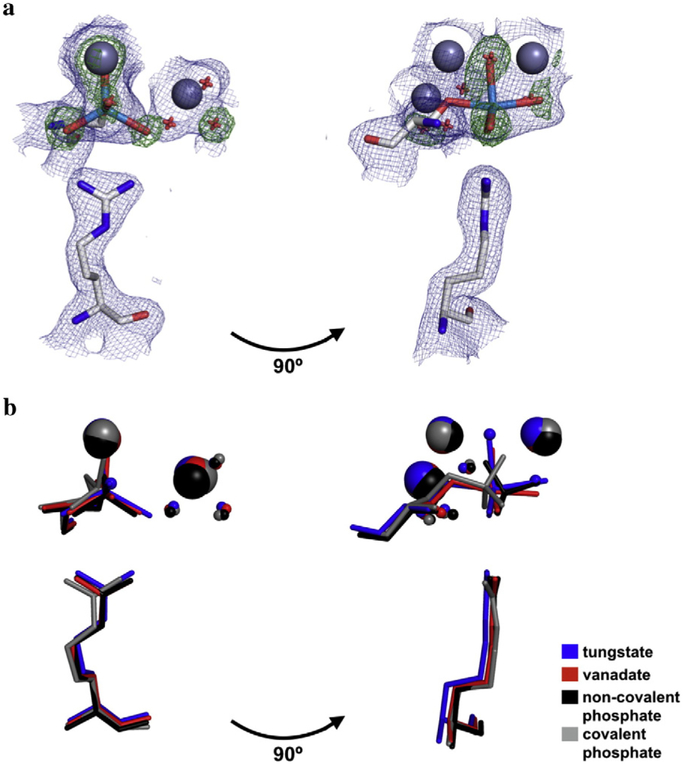
Fig. 2.
Structure of the tungstate-bound AP active site and in comparison to other bound anionic ligands. (a) Overlay of maps of the 2Fo-Fc electron density (contoured at 1σ) of the final model (blue) and the Fo-Fc map electron density (contoured at 4σ) from the simulated annealing omit map in which the oxygen atoms of the tungstate ion were omitted (green). Zn2+ ions are colored dark gray; tungstate, light blue; Ser102 and Arg166, light gray; waters, red. For clarity, only one active site is shown. (b) Crystal structures of AP bound to tungstate, vanadate, and phosphate (both covalently and non-covalently) were overlaid to compare the positions of ligands with respect to Ser102, Arg166, the active site Zn2+ and Mg2+ ions, and the Mg2+-coordinated water molecules. In the tungstate structure, the ligand was modeled in at partial occupancy, with partially occupied water molecules at sites that presumably coordinate the Zn2+ ions in the fraction of enzyme without bound tungstate. The PDB codes for vanadate, non-covalently bound phosphate, and covalently bound phosphate are 1B8J, 3TG0, and 1HJK, respectively.