Fig. 5.
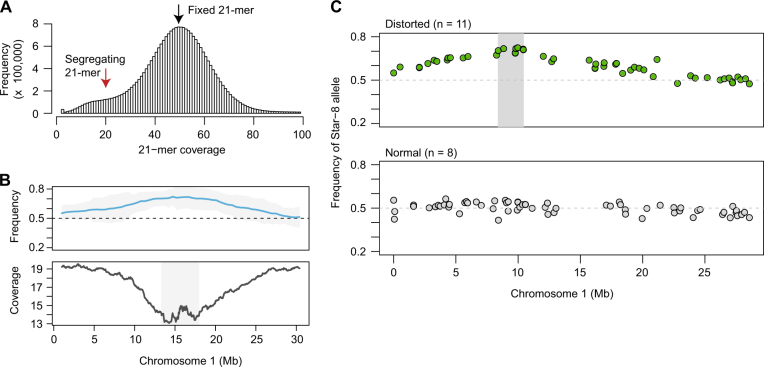
Mapping intervals refined using k-mer coverage and bulked segregant analysis. a The coverage of unique 21 nt k-mers is plotted for POP035 (ICE63 × Vash-1) after whole-genome resequencing. The first peak in coverage represents 21-mers found in only one of the two grandparents (red arrow), while the second, larger peak represents those sequences found in both (black arrow). b The upper panel displays the beta-binomial modeled allele frequency estimates (blue) and their 95% confidence intervals (grey) for POP035 as described in the legend for Fig. 2. In the lower panel, the coverage of 21-mers unique to only one of the two grandparents (coverage < 25×) is plotted in 1 Mb sliding windows (50 kb steps). Coverage decreases in the candidate regions. Intervals (grey box) are defined by merging windows with values within 1× coverage of the minimal window in each population. c Bulked segregant analysis was performed for Star-8, an accession that repeatedly contributed distorted loci. Sequencing reads were combined for populations exhibiting distortion when crossed with Star-8, and for populations not exhibiting distortion when crossed to Star-8 (normal pool). A candidate interval (grey box) was obtained by merging all segregating positions within 5% of the maximal allele frequency