Extended Data Figure 2.
Features of ultrahigh-depth targeted amplicon sequencing used for validation.
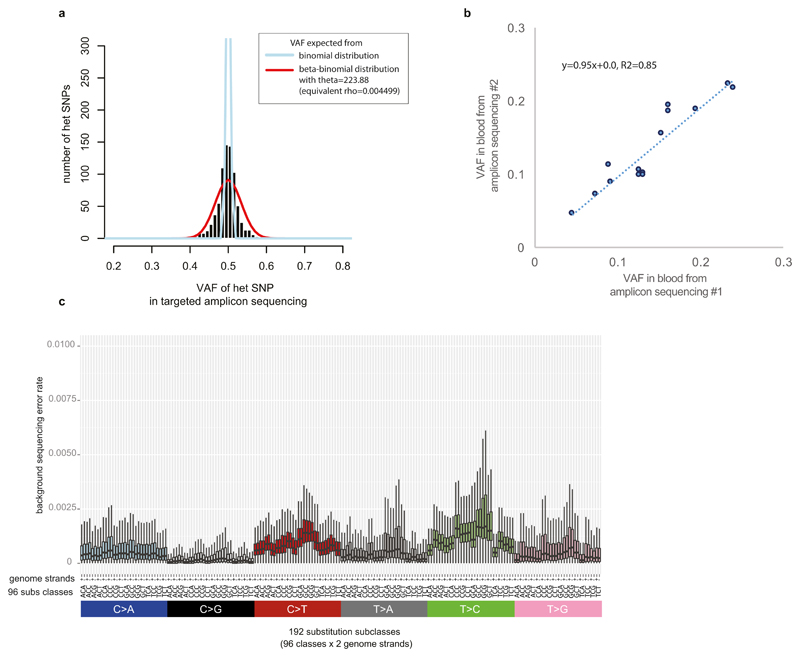
a, Estimation of the impact of potential PCR allelic bias from targeted amplicon sequencing. Using inherited heterozygous SNP sites which were PCR amplified and ultra-deep sequenced, we assessed potential PCR bias (i.e. preferential amplification of one allele compared to the other): the distribution of VAFs was broader than expected from a binomial distribution (theoretical maximum), but the PCR bias was not substantial as a clear peak at VAF=0.5 was present. The estimated overdispersion level (theta value in beta-binomial distribution) was 223.88. The estimate was used in the simulation studies for assessment of cell doubling asymmetry in early embryogenesis (see Methods for more details).
b, High precision of ultrahigh-depth amplicon sequencing in assessment of VAF of a mutation. For the 14 early embryonic mutations, we quantified their VAFs from the second blood samples using the same strategy (i.e. PCR amplification and deep sequencing). The VAF estimates from the first and the second sequencings were highly correlated.
c, Background error rate of targeted amplicon sequencing (see Methods). The background mutation rate showed sequence context dependency. Error bars denote 2 * interquartile range. We used these background mutation rates in a filtering step.