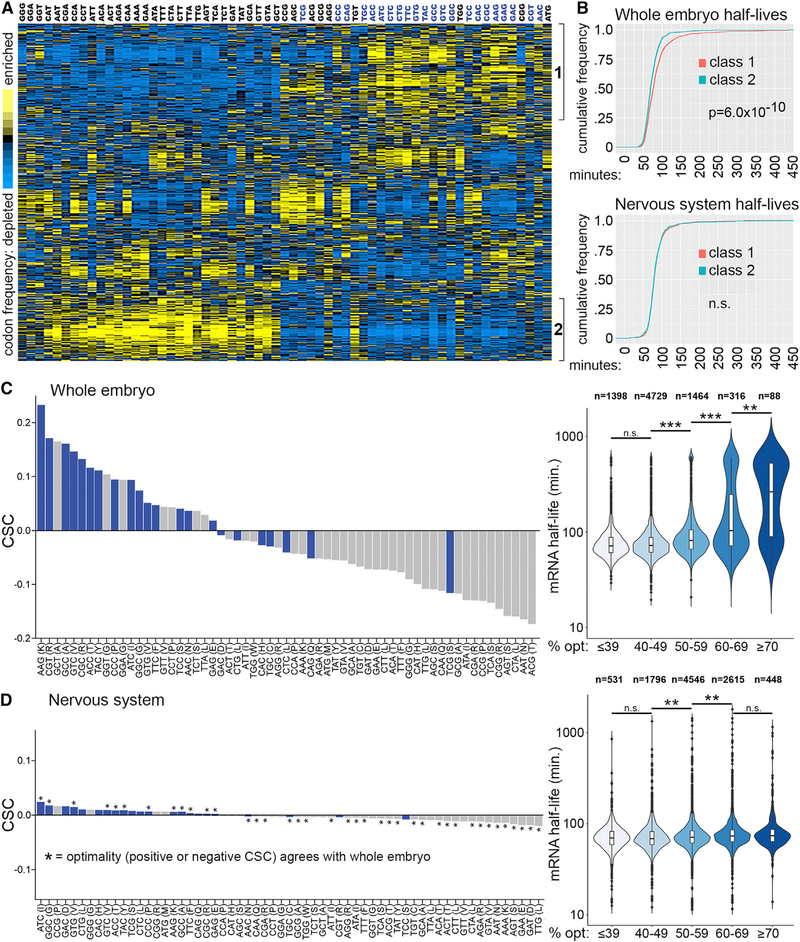
Figure 1. Codon Usage Correlates with Embryo mRNA Stability but Not Neural mRNA Stability.
(A) 3,312 mRNAs clustered according to codon usage. Each row is a different mRNA and each column represents the frequency of the codon listed on top.Codons in blue type are predicted preferred codons.
(B) Distribution of mRNA half-lives for class 1 versus class 2 mRNAs based on whole embryo decay measurements (top panel) and neural-specific decay measurements (bottom panel). p value determined by Kolmogorov-Smirnov test, n.s., no significant difference between classes.
(C) Codon stabilization coefficients (left) and mRNA half-lives categorized by percent optimal codon content (right) based on whole embryo measurements(7,995 mRNAs).
(D) Codon stabilization coefficients for all codons (left) and mRNA half-lives categorized by optimal codon content (right) based on neural-specific measurements (9,699 mRNAs). Codons with the same CSC direction (positive or negative) in the whole embryo and neural-specific datasets are indicted with an asterisk. Significant differences among categories in the violin plots of (C) and (D) were detected by Kruskal-Wallis test and p values (***p < 1 × 1010, **p < 0.001, or no significant difference [n.s.]) for the indicated pairwise comparisons are based on Dunn’s test. See also Table S1.