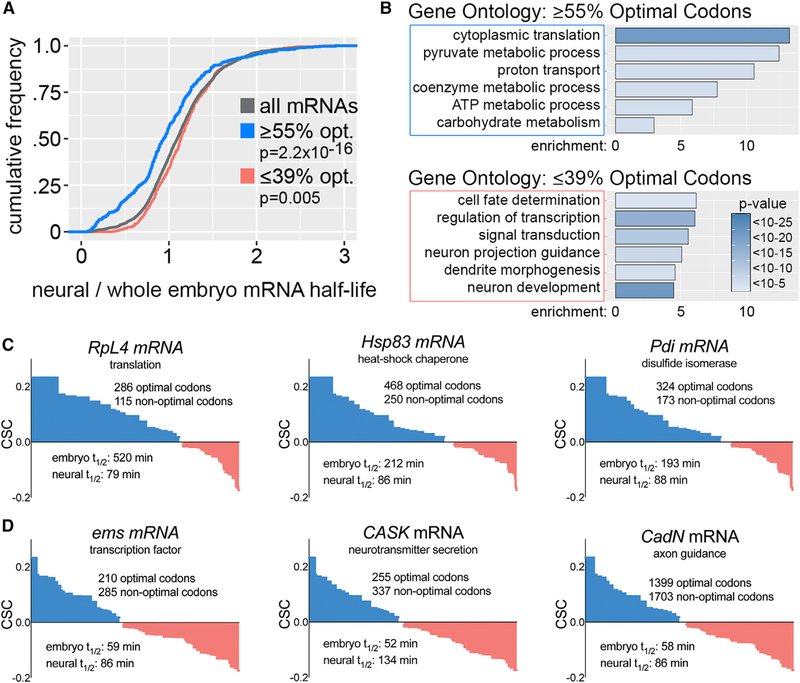
Figure 2. Codon Optimality Establishes Decay of Functionally Related Transcripts.
(A) Relative stability of mRNAs based on neural-specific versus whole embryo measurements. x axis is the neural-specific half-life divided by whole embryohalf-life. p values are based on Kolmogorov-Smirnov test comparing all mRNAs to high optimality or low optimality mRNAs.
(B) Gene ontology categories enriched in high optimality versus low optimality mRNAs.
(C) Codon content and differential stability of representative high optimality mRNAs. The codon content of each gene is plotted in order of optimality (high to low) as defined by whole embryo CSCs.
(D) Codon content and differential stability of representative low optimality mRNAs. Codon content is plotted as in (C). See also Table S1.