In this study, Horie et al. used whole-embryo single-cell RNA-seq assays to identify the regulatory determinants responsible for the specification of dopaminergic neurons in the CNS of the Ciona tadpole. Their findings suggest that the reiterative use of functional manipulations and single-cell RNA-seq assays is an effective means for the identification of regulatory cocktails underlying the specification of specific cell identities.
Keywords: ascidian, Ciona CNS, dopamine, neuronal differentiation, single-cell transcriptomics
Abstract
The CNS of the protovertebrate Ciona intestinalis contains a single cluster of dopaminergic (DA) neurons, the coronet cells, which have been likened to the hypothalamus of vertebrates. Whole-embryo single-cell RNA sequencing (RNA-seq) assays identified Ptf1a as the most strongly expressed cell-specific transcription factor (TF) in DA/coronet cells. Knockdown of Ptf1a activity results in their loss, while misexpression results in the appearance of supernumerary DA/coronet cells. Photoreceptor cells and ependymal cells are the most susceptible to transformation, and both cell types express high levels of Meis. Coexpression of both Ptf1a and Meis caused the wholesale transformation of the entire CNS into DA/coronet cells. We therefore suggest that the reiterative use of functional manipulations and single-cell RNA-seq assays is an effective means for the identification of regulatory cocktails underlying the specification of specific cell identities.
Dopaminergic (DA) neurons mediate a variety of “reward” behaviors in vertebrates, such as feeding, mating, and response to external stimuli (Iversen and Iversen 2007). There are several classes of DA neurons in the vertebrate CNS, including those mediating motor functions in the midbrain and others controlling secretion of neuropeptides in the hypothalamus (Flames and Hobert 2011; Grattan 2015). There is considerable information about the specification of midbrain DA neurons due to their importance in Parkinson's disease (Flames and Hobert 2011; Kee et al. 2017; Kirkeby et al. 2017; Parmar 2018). Characterization of the molecular programs controlling differentiation of DA neurons is crucial for understanding this important neuronal cell type and developing stem cell-based therapies for DA deficiencies such as Parkinson's disease (Parmar 2018).
Previous studies identified the Ets transcription factor (TF) AST-1 as a key determinant of DA neurons in the nematode worm Caenorhabditis elegans (Flames and Hobert 2009; Doitsidou et al. 2013). This mechanism is conserved in olfactory bulb DA neurons in vertebrates (Flames and Hobert 2009; Doitsidou et al. 2013). However, vertebrates possess additional classes of DA neurons, and additional studies are required to identity corresponding mechanisms. Several TFs have been implicated in the differentiation of midbrain DA neurons in vertebrates, including Nurr, Lmx, and Pitx3 (Flames and Hobert 2011). Considerably less is known about other classes of DA neurons, such as those associated with the hypothalamus. For this purpose, we sought to identify selector genes for DA neurons in the protovertebrate Ciona intestinalis.
The CNS of C. intestinalis consists of 177 neurons that share a number of similarities with the vertebrate CNS (Ryan et al. 2016). The tadpole contains a single cluster of DA neurons, the coronet cells, which are located in ventral regions of the sensory vesicle in close proximity to group III photoreceptor cells (Fig. 1A; Moret et al. 2005a,b; Horie et al. 2008; Razy-Krajka et al. 2012; Ryan et al. 2016; Sharma et al. 2018). This region of the ascidian CNS shares a number of similarities with the hypothalamus (Moret et al. 2005a,b; Razy-Krajka et al. 2012) and is evocative of the deep brain light sensory system of lower fish (Nakane et al. 2013). Here we used whole-embryo single-cell RNA sequencing (RNA-seq) assays to elucidate the regulatory networks underlying the specification of DA neurons/coronet cells in the Ciona tadpole.
Figure 1.
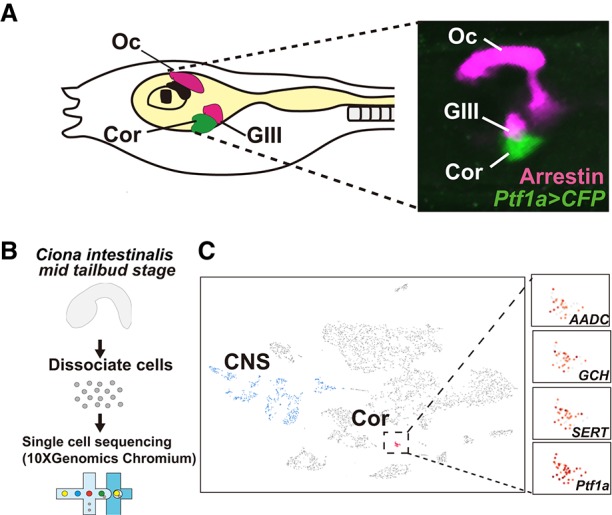
Whole-embryo single-cell RNA-seq analysis of coronet cells. (A, left) Diagram of a Ciona tadpole showing the position of coronet cells (DA neurons [green]) and photoreceptor cells, including the ocellus and group III cells (magenta). (Right) Coronet cells visualized by a Ptf1a>CFP reporter gene (green) containing 5′ flanking regulatory sequences from Ptf1a and photoreceptor cells (magenta) visualized by immunostaining with an Arrestin antibody. (Oc) Ocellus; (Cor) coronet cells; (GIII) group III photoreceptor cells. (B) Schematic illustrating the workflow for single-cell RNA-seq analysis of Ciona embryos using the 10x Genomics Chromium system. (C) A t-distributed stochastic neighbor embedding (tSNE) projection map of mid-tail bud stage embryos highlighting the distribution of DA neuron marker genes (AADC, GCH, and SERT) and Ptf1a. Each dot corresponds to the transcriptome of a single cell. Red dots indicate DA neurons/coronet cells clusters, blue dots indicate CNS, and gray dots indicate other tissues.
Results and Discussion
Mid-tail bud embryos (∼1500 cells) were dissociated and barcoded using the 10x Genomics Chromium system (Fig. 1B). RNA was extracted from individual cells and reverse-transcribed, and the resulting cDNAs were sequenced. A total of ∼5000 cells was sequenced to ensure effective coverage of the entire embryo. The single-cell transcriptome profiles identified all of the major tissues, including notochord, endoderm, mesenchyme, tail muscles, and CNS (Fig. 1C; Horie et al. 2018). DA neurons/coronet cells were identified on the basis of their expression of a variety of dopaminergic markers genes, including those encoding the dopamine biosynthetic pathway such as TH, GCH, and AADC (Fig. 1C; Supplemental Figs. S1, S2; Supplemental Table 1). These cells form a discrete cluster that is distinct from all other neuronal cell types in the CNS (Fig. 1C, red dots).
The 10x analysis also identified a number of transcripts in DA neurons/coronet cells that encode secreted neuropeptides (Supplemental Fig. S2; Hamada et al. 2011; Kawada et al. 2011). This observation supports and extends previous proposals that coronet cells are a component of an ancient protohypothalamic–retinal territory (Fig. 1A; Razy-Krajka et al. 2012). In vertebrates, there are separate DA neurons and neurosecretory neurons (Grattan 2015). In contrast, both activities are contained within individual coronet cells, raising the possibility that cellular subfunctionalization contributed to the evolution of the hypothalamus (Arendt 2008).
The transcriptome profiles of DA neurons/coronet cells identified a number of regulatory genes, including Ptf1a, a basic helix–loop–helix (bHLH) gene implicated in the development of the pancreas and GABAergic/glutamatergic neurons in the cerebellum of vertebrates (Hoshino et al. 2005; Fujitani et al. 2006; Dullin et al. 2007; Nakhai et al. 2007). It is specifically expressed in DA neurons/coronet cells and absent in all other major neuronal cell types in mid-tail bud embryos (Fig. 1C; Supplemental Figs. S1, S2; Razy-Krajka et al. 2012). To determine the role of Ptf1a in the specification of DA neurons/coronet cells, we inhibited gene activity by injecting a Ptf1a morpholino (MO) that targets the 5′ untranslated region (Supplemental Fig. S3). The resulting morphants appear normal, although DA marker gene expression is lost in ventral regions of the sensory vesicle (Fig. 2B, cf, with A; Supplemental Fig. S4). There is a corresponding expansion in the expression of arrestin, raising the possibility that DA neurons/coronet cells are transformed into photoreceptor cells in Ptf1a morphants (Supplemental Fig. S5). These results suggest that Ptf1a is important for the development of DA neurons/coronet cells in the Ciona CNS.
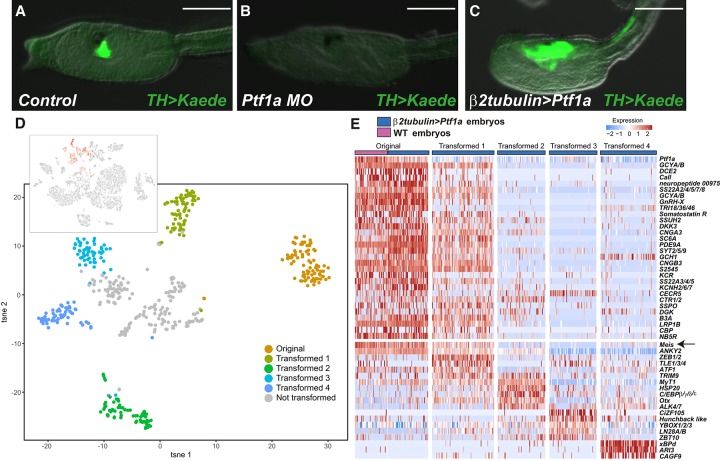
Figure 2.
Ptf1a is required for the differentiation of DA/coronet cells. (A–C) Head regions of TH>Kaede transgenic larvae. (A) Kaede expression in the coronet cells of control larvae (51 of 103 larvae displayed this expression pattern). (Note that the transgenic line is a heterozygote for the TH>Kaede transgene.) (B) Same as A except that it was injected with a Ptf1a MO (111 of 121 larvae displayed this expression pattern) (see Supplemental Fig. S3 for more details). (C) Same as A except that Ptf1a was misexpressed throughout the CNS by β2tubulin 5′ regulatory sequences (29 of 60 larvae displayed this expression pattern). Bar, 100 µm. (D, top) The tSNE projection map of mid-tail bud embryos expressing the β2tubulin>Ptf1a transgene. Red dots identify cells expressing a β2tubulin>CFP reporter gene. (Bottom) tSNE “subclustering” of cells expressing Ptf1a and the CFP marker gene. (E) Heat map of native DA neurons/coronet cells and four different groups of transformed cells (shown in D) showing the relative expression of a select group of genes encoding cellular effectors and TFs. Gene expression profiles of transformed cells in cluster 1 are similar to those of native DA neurons/coronet cells. The arrow identifies Meis, which is expressed in native DA neurons/coronet cells and transformed cluster 1 but not the partially transformed cells in clusters 2, 3, or 4.
To determine whether Ptf1a functions as a “master control gene” for DA neuron/coronet cell identity, we misexpressed it throughout the nervous system (CNS and peripheral nervous system [PNS]) using 5′ regulatory sequences from the β2tubulin gene. Injection of the β2tubulin>Ptf1a transgene resulted in expanded expression of the TH>Kaede marker gene throughout the sensory vesicle and portions of the nerve cord (Fig. 2C, cf. with A). This result strengthens the evidence that Ptf1a functions as a determinant of DA neurons/coronet cells. It would appear that misexpression of Ptf1a is sufficient to transform some regions of the CNS into DA neurons/coronet cells but not others.
To explore the nature of the transformations, we performed single-cell RNA-seq assays with transgenic embryos expressing the β2tubulin>Ptf1a transgene along with a β2tubulin>CFP reporter gene that identifies all of the cells expressing Ptf1a (Fig. 2D; Supplemental Fig. S6). Most tissues do not show any changes in gene activity (e.g., notochord, endoderm, and tail muscle) (Supplemental Fig. S6). In contrast, the CNS and PNS display strong expression of Ptf1a (Fig. 2D, top panel, Supplemental Fig. S7A). Ptf1a-expressing cells were clustered based on their transcriptome profiles (Fig. 2D, bottom panel; Supplemental Fig. S7A; Satija et al. 2015; Butler et al. 2018). Cluster 1 displays the most complete transformation, since it expresses nearly the complete suite of DA neurons/coronet identity genes (Fig. 2E, heat map; Supplemental Figs. S6, S7B). It is possible that these cells derive from posterior regions of the sensory vesicle and nerve cord (Fig. 2C). In contrast, clusters 2–4 display progressively fewer DA neurons/coronet marker genes and continue to express a variety of marker genes reflecting their origins from anterior regions of the sensory vesicle and PNS.
Misexpression of Ptf1a suppresses the development of photoreceptor cells (Horie et al. 2008) and ependymal cells (Supplemental Fig. S5; Horie et al. 2011) but does not significantly alter glutamatergic, GABAergic/glycinergic, and cholinergic neurons (Supplemental Fig. S8). The loss of photoreceptor cells and ependymal cells suggests that they are transformed into supernumerary DA neurons/coronet cells upon misexpression of Ptf1a. It seems likely that Ptf1a works with additional sequence-specific TFs to specify DA neurons/coronet cells. These factors may be present in photoreceptor cells and ependymal cells but absent or only weakly expressed in other neurons. We found that the homeobox gene Meis fulfills these criteria (Fig. 2E; Supplemental Fig. S9). Previous studies have shown that Meis functions cooperatively with a number of Hox TFs (Moens and Selleri 2006; Agoston et al. 2014). To determine whether it might also work in concert with Ptf1a to specify DA neurons/coronet cells, we simultaneously misexpressed both genes (Fig. 3).
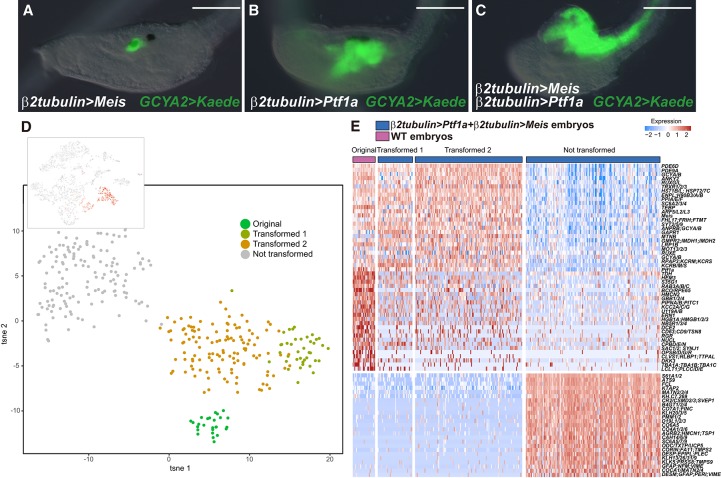
Figure 3.
A Ptf1a + Meis cocktail promotes the differentiation of DA neurons/coronet cells. (A–C) Head regions of larvae that were injected with a GCYA2>Kaede reporter gene that is specifically expressed in DA neurons/coronet cells (see Supplemental Fig. S2). (A) The embryo was coinjected with a β2tubulin>Meis transgene. Misexpression of Meis does not alter the normal expression of the reporter gene within DA neurons/coronet cells (100 of 100 larvae displayed this expression pattern). (B) The embryo was coinjected with the β2tubulin>Ptf1a transgene. Kaede expression is expanded into posterior regions of the sensory vesicle and anterior neural tube (104 of 104 larvae displayed this expression pattern) (see Fig. 2C). (C) The embryo was coinjected with both β2tubulin>Ptf1a and β2tubulin>Meis transgenes. The GCYA2 reporter gene is now expressed throughout the entire CNS (89 of 89 larvae displayed this expression pattern). Bar, 100 µm. (D, top) A tSNE projection map of late tail bud stage embryos expressing both the Ptf1a and Meis transgenes. Red dots correspond to cells expressing the β2tubulin>CFP reporter gene, which identifies cells that misexpress Ptf1a and Meis. (Bottom) tSNE subclustering of coexpressing cells. (E) Heat map of native DA neurons/coronet cells, transformed cluster 1, transformed cluster 2, and untransformed cells. A select group of genes encoding cellular effectors and TFs is shown. Clusters 1 and 2 display transcriptome profiles that are very similar to those seen for native coronet cells. The untransformed cells are likely to correspond to epidermis based on their transcriptome profiles.
β2tubulin>Ptf1a and β2tubulin>Meis transgenes were coinjected in unfertilized eggs and grown to the late tail bud I (LTB I) stage. The resulting embryos exhibit a dramatic transformation of the entire CNS into DA neurons/coronet cells (Fig. 3C, cf. A and B). To determine the nature of this transformation, we performed single-cell RNA-seq assays on transformed embryos (Fig. 3D,E; Supplemental Figs. S10, S11). Most of the transformed cells express the complete suite of DA neuron/coronet cell marker genes. They lack expression of marker genes identifying their developmental origins and original neuronal identities, such as VACHT/ChAT (cholinergic neurons) and GAD (GABAergic neurons) (Fig. 3E; Supplemental Fig. S12). In contrast, the other major site of expression mediated by β2tubulin regulatory sequences—epidermal sensory neurons—displays little or no transformation toward a DA neuron/coronet cell identity.
It is likely that Ptf1a and Meis work directly to regulate target genes that are specifically expressed in DA neurons/coronet cells. The 5′ flanking regions of many such genes contain tightly linked Ptf1a-binding (E box) and Meis-binding sites (Agoston et al. 2014), and DNA fragments containing these motifs mediate restricted expression in DA neurons/coronet cells when attached to reporter genes and expressed in transgenic embryos (Fig. 4; Supplemental Figs. S13–S15). Given the parallels between DA neurons/coronet cells and the hypothalamus, it seems reasonable to suggest that the regulatory “cocktail” of Ptf1a and Meis might also control the development of DA neuronal cell types in vertebrates. Indeed, Ptf1a has been suggested to play a role in the specification of DA neurons in the hypothalamus in mice (Fujiyama et al. 2018), while Meis TFs have been implicated in the specification of DA neurons in the olfactory bulb (Agoston et al. 2014) and hypothalamus (Hook et al. 2018) in mice.
Figure 4.
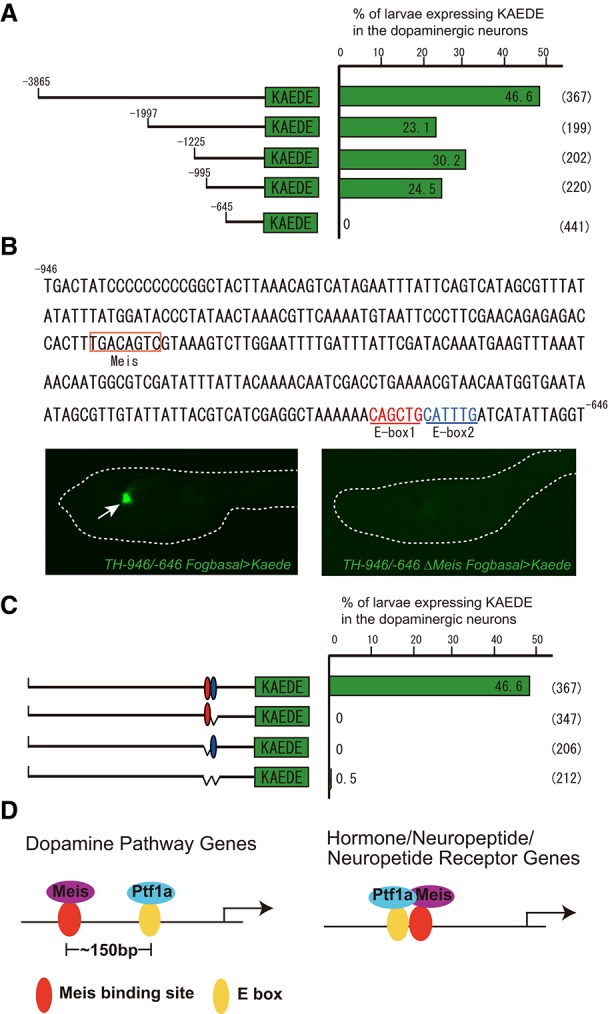
Gene network for the specification of DA neurons/coronet cells. (A) Deletion analyses of the cis regulatory region of Ci-TH. An ∼350-base-pair (bp) DNA fragment (−995 to −646 bp upstream of the translation start site) from the 5′ flanking region of Ci-TH is sufficient to mediate Kaede expression in DA neurons/coronet cells. (B) This 5′ regulatory DNA contains two E-box sequences (E-box1 and E-box2) and one Meis-binding site. Deletion in the Meis-binding site eliminates expression in DA neurons/coronet cells (116 of 119 larvae displayed this pattern). (C) Deletion and mutation analysis of the E-box sequences. Deletion and mutation analysis of E-box1 and E-box2 showed that binding sites are necessary for Ci-TH expression in DA neurons/coronet cells. (D) Schematic diagrams of enhancer regions mediating localized expression in DA neurons/coronet cells. (Left) Three of three DA neuron marker genes contain enhancers with distant linkage of Meis- and Ptf1a-binding motifs (∼150 bp). (Right) In contrast, two of two neuroendocrine genes contain enhancers with tightly linked Meis- and Ptf1a-binding motifs.
Single-cell technologies are pervasively used to provide descriptive cell atlases of gene expression (Karaiskos et al. 2017; Briggs et al. 2018; Farrell et al. 2018; Fincher et al. 2018; Plass et al. 2018; Wagner et al. 2018). Here, we attempted to extend the promise of these technologies by combining them with classical approaches in experimental embryology. We used whole-embryo single-cell transcriptome assays to identify the determinants of DA neurons/coronet cells in the Ciona larval CNS. The identification of Meis as a critical cofactor of Ptf1a in the specification of DA neurons/coronet cells depended on single-cell analysis. It is broadly distributed in the CNS, PNS, and other tissues but preferentially expressed in those regions of the CNS most susceptible to transformation by Ptf1a (e.g., photoreceptor cells and ependymal cells). Previous studies have focused on individual determinants, although these are not always sufficient to specify specific cell types (Flames and Hobert 2009; Doitsidou et al. 2013). The reiterative use of experimental manipulations (e.g., misexpression of Ptf1a) and single-cell analysis provides a potent one–two punch in the identification of the complex gene networks underlying development.
Materials and methods
Biological materials
Wild-type C. intestinalis type A (also called Ciona robusta) adults were obtained from M-Rep and the National Bio-Resource Project for Ciona in Japan. Sperm and eggs were collected by dissecting the sperm and gonadal ducts. Transgenic lines were cultured and maintained in an island system.
Isolation and characterization of Ci-Ptf1a cDNA
A partial cDNA fragment of Ci-Ptf1a was found in the Ghost database (http://ghost.zool.kyoto-u.ac.jp/cgi-bin/gb2/gbrowse/kh). To obtain the full-length coding sequence, we performed 5′ RACE using Generacer kit (Invitrogen). The nucleotide sequences of oligonucleotide primers used for 5′ RACE were 5′-CACCACCCCTTCTTCGGTAAATTGGAAG-3′ (for the primary PCR) and 5′-TCGGGAGGCTAGTACCTCACGAAGCAACG-3′ (for the nested PCR). The cDNA fragments were cloned into a pGemT vector (Promega). The cDNA clone was sequenced on both strands with automatic DNA sequencer (Applied Biosystems).
Whole-mount in situ hybridization
A cDNA clone of Ci-Ptf1a that contained the full ORF was used as the template to synthesize a digoxigenin-labeled antisense RNA probe using a DIG-RNA labeling kit (Roche). In situ hybridization of the whole-mount specimens was carried out as described previously (Kusakabe et al. 2002).
Constructs
Reporter genes were designed using previously published enhancer sequences TH, AADC, GCH, SERT, and Ptf1a (Razy-Krajka et al. 2012). To generate pSPCiPtf1a ΔMO target sequence C, 5′ upstream regions of Ci-Ptf1a were amplified by PCR using a thermostable DNA polymerase (PrimeSTAR HS DNA polymerase, Takara) and oligonucleotide primers (Supplemental Table 2). The PCR products were digested with BamHI and inserted into the BamHI pSPeCFP. To generate pSPCiGCYA2K, pSPCiGCYBK, pSPCiGnRHXK, pSPCiNtlBK, pSPCiPDE9aK, pSPPDEdK, pSPLectinK, and pSPSS23A3K, 5′ upstream regions of Ci-GCYA2 Ci-GCYB, Ci-GnRHX, Ci-NtlB, Ci-PDE9a, Ci-PDEd, Ci-Lectin, and Ci-SS23A3 were amplified by PCR using a thermostable DNA polymerase and oligonucleotide primers (Supplemental Table 2). The PCR products were digested with BamHI/NotI and inserted into the BamHI/NotI site of pSPKaede (Hozumi et al. 2010).
To generate pMiCiTHK, a promoter-Kaede cassette was amplified by PCR using a thermostable DNA polymerase and vector-specific oligonucleotide primers (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAACTCGAGCAGCTGAAGCTTG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGCAGATCTGATGGCCGCTTTGAC-3′). The PCR product was subcloned into pMiDestF (Sasakura et al. 2008) with a gateway system (Invitrogen). To generate pSPCiPtf1acDNA, the coding sequence of Ci-Ptf1a was amplified by PCR with oligonucleotide primers (Supplemental Table 2). The PCR product was digested with NotI and inserted into the NotI and blunted EcoRI sites of pSPeGFP. To generate pSPCiMeiscDNA, the coding sequence of Ci-Meis was amplified by PCR with oligonucleotide primers (Supplemental Table 2). The PCR product was digested with BamHI/EcoRV and inserted into the BamHI and blunted EcoRI sites of pSPeGFP.
To generate the pSPCiβ2tubulinCiPtf1acDNA and pSPCiPtf1a ΔMO target sequence Ptf1acDNA, 5′ upstream regions of Ci-β2tubulin and Ci-Ptf1a ΔMO target sequences were inserted into the BamHI site of pSPCiPtf1acDNA. To generate pSPCiβ2tubulinCiMeisacDNA, 5′ upstream regions of Ci-β2tubulin were inserted into the XhoI and BamHI sites of pSPCiMeiscDNA.
Generation of tranagenic lines
TH>Kaede transgenic lines were created by coelectroporation of in vitro synthesized transposase mRNA and pMiCiTHK (Sasakura 2007). Electroporated animals were cultured and maintained in an island system. Screening of transgenic lines was performed as described previously (Sasakura et al. 2007).
Microinjection of antisense MO oligonucleotides
MO oligonucleotides were obtained from Gene Tools, LLC. The antisense oligonucleotide sequence of the MO against Ci-Ptf1a was 5′-CGTTGATAACTCACAAACACATAGG-3′. MOs were dissolved in DEPC-treated water containing 1 mg/mL tetramethylrhodamine dextran (Invitrogen, D1817). The concentrations of MO and plasmid DNA in the injection medium were 0.5 mM and 2.5–10 ng/µL, respectively. Microinjections of MOs and reporter constructs were performed as described previously (Horie et al. 2018). All experiments were repeated at least twice with different batches of embryos.
Single-cell RNA-seq assays
Eggs injected with 2.5 ng/µL β2tubulin>Ptf1a or 2.5 ng/µL β2tubulin>Ptf1a + 2.5 ng/µL β2tubulin>Meis and control eggs were fertilized side by side and allowed to develop to the middle or LTB stage (11 h or 13.5 h after fertilization at 18°C). For each sample, 120 morphologically normal embryos were used for single-cell RNA-seq assays. Dissociation of the embryos and single-cell RNA-seq assays by the 10x Genomics Chromium system were done as described previously (Horie et al. 2018) and are detailed in the Supplemental Material.
Image acquisition
Images of transgenic larvae were obtained with a Zeiss AxioPlan, Zeiss AX 10 epifluorescence microscope, and Olympus Fluoview FV10i confocal microscope.
Accession number
10x single-cell RNA-seq data for Ptf1a, Ptf1a and Meis overexpression, and control are available from Gene Expression Omnibus (GEO) under the accession number GSE120035.
Supplementary Material
Acknowledgments
We thank all the members of the Lewis-Sigler Institute Genome Facility for technical support of the single-cell RNA-seq assays and analysis. We also thank Reiko Yoshida, Chikako Imaizumi, and all other members of the staff at the Maizuru Fisheries Research Station of Kyoto University for providing C. intestinalis. This study was supported by a grant from the National Institutes of Health to M.L. (NS076542) and Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to Y.S. (20681019 and 16H04815), T.H. (24687008 and 16K07433), T.G.K. (16H04724), and N.S. (20247031 and 16H04824). T.H. was supported by Pre-Strategic Research for Embryonic Science and Technology from the Japan Science and Technology Agency and by Pre-Strategic Initiatives from University of Tsukuba. Y.S. was supported by the Toray Science and Technology Grant. This study was further supported by the National Bio-Resource Project (NBRP) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Author contributions: T.H., Y.S., and M.L. conceived the project and designed the experiments. T.H., R.H., and K.C. performed the experiments. T.G.K and M.N. provided essential materials. T.H., R.H., K.C., C.C., Y.S., and M.L. analyzed and interpreted the data. T.H., N.S., and Y.S. wrote the first draft. T.H., K.C., and M.L. wrote the final manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.317669.118.
References
- Agoston Z, Heine P, Brill MS, Grebbin BM, Hau AC, Kallenborn-Gerhardt W, Schramm J, Götz M, Schulte D. 2014. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141: 28–38. [DOI] [PubMed] [Google Scholar]
- Arendt D. 2008. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet 9: 868–882. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM. 2018. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360: eaar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Flames N, Topalidou I, Abe N, Felton T, Remesal L, Popovitchenko T, Mann R, Chalfie M, Hobert O. 2013. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev 27: 1391–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dullin JP, Locker M, Robach M, Henningfeld KA, Parain K, Afelik S, Pieler T, Perron M. 2007. Ptf1a triggers GABAergic neuronal cell fates in the retina. BMC Dev Biol 7: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. 2018. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360: eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW. 2018. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360: eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Hobert O. 2009. Gene regulatory logic of dopamine neuron differentiation. Nature 458: 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Hobert O. 2011. Transcriptional control of the terminal fate of monoaminergic neurons. Annu Rev Neurosci 34: 153–184. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, et al. 2006. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Develoment 133: 4439–4450. [DOI] [PubMed] [Google Scholar]
- Fujiyama T, Miyashita S, Tsuneoka Y, Kanemaru K, Kakizaki M, Kanno S, Ishikawa Y, Yamashita M, Owa T, Nagaoka M, et al. 2018. Forebrain Ptf1a is required for sexual differentiation of the brain. Cell Rep 24: 79–94. [DOI] [PubMed] [Google Scholar]
- Grattan DR. 2015. 60 Years of neuroendocrinology: the hypothalamo–prolactin axis. J Endocrinol 226: T101–T122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Shimozono N, Ohta N, Satou Y, Horie T, Kawada T, Satake H, Sasakura Y, Satoh N. 2011. Expression of neuropeptide- and hormone-encoding genes in the Ciona intestinalis larval brain. Dev Biol 352: 202–214. [DOI] [PubMed] [Google Scholar]
- Hook PW, McClymont SA, Cannon GH, Law WD, Morton AJ, Goff LA, McCallion AS. 2018. Single-cell RNA-seq of mouse dopaminergic neurons informs candidate gene selection for sporadic Parkinson disease. Am J Hum Genet 102: 427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Sakurai D, Ohtsuki H, Terakita A, Shichida Y, Usukura J, Kusakabe T, Tsuda M. 2008. Pigmented and nonpigmented ocelli in the brain vesicle of the ascidian larva. J Comp Neurol 509: 88–102. [DOI] [PubMed] [Google Scholar]
- Horie T, Shinki R, Ogura Y, Kusakabe TG, Satoh N, Sasakura Y. 2011. Ependymal cells of chordate larvae are stem like cells that form the adult nervous system. Nature 469: 525–528. [DOI] [PubMed] [Google Scholar]
- Horie R, Hazbun A, Chen K, Cao C, Levine M, Horie T. 2018. Shared evolutionary origin of vertebrate neural crest and cranial placodes. Nature 560: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, et al. 2005. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47: 201–213. [DOI] [PubMed] [Google Scholar]
- Hozumi A, Kawai N, Yoshida R, Ogura Y, Ohta N, Satake H, Satoh N, Sasakura Y. 2010. Efficient transposition of a single Minos transposon copy in the genome of the ascidian Ciona intestinalis with a transgenic line expressing transposase in eggs. Dev Dyn 239: 1076–1088. [DOI] [PubMed] [Google Scholar]
- Iversen SD, Iversen LL. 2007. Dopamine: 50 years in perspective. Trends Neurosci 30: 188–193. [DOI] [PubMed] [Google Scholar]
- Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, Kipar C, Kocks C, Rajewsky N, Zinzen RP. 2017. The Drosophila embryo at single-cell transcriptome resolution. Science 358: 194–199. [DOI] [PubMed] [Google Scholar]
- Kawada T, Ogasawara M, Sekiguchi T, Aoyama M, Hotta K, Oka K, Satake H. 2011. Peptidomic analysis of the central nervous system of the protochordate, Ciona intestinalis: homologs and prototypes of vertebrate peptides and novel peptides. Endocrinology 152: 2416–2427. [DOI] [PubMed] [Google Scholar]
- Kee N, Volakakis N, Kirkeby A, Dahl L, Storvall H, Nolbrant S, Lahti L, Björklund AK, Gillberg L, Joodmardi E, et al. 2017. Single-cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell 20: 29–40. [DOI] [PubMed] [Google Scholar]
- Kirkeby A, Nolbrant S, Tiklova K, Heuer A, Kee N, Cardoso T, Ottosson DR, Lelos MJ, Rifes P, Dunnett SB, et al. 2017. Predictive markers guide differentiation to improve graft outcome in clinical translation of hESC-based therapy for Parkinson's disease. Cell Stem Cell 20: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T, Ysohida R, Kawakami I, Kusakabe R, Mochizuki Y, Yamada L, Shin-I T, Satoh N, Tsuda M, et al. 2002. Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev Biol 242: 188–203. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. 2006. Hox cofactors in vertebrate development. Dev Biol 291: 193–206. [DOI] [PubMed] [Google Scholar]
- Moret F, Christiaen L, Deyts C, Blin M, Joly JS, Vernier P. 2005a. The dopamine-synthesizing cells in the swimming larva of the tunicate Ciona intestinalis are located only in the hypothalamus-related domain of the sensory vesicle. Eur J Neurosci 21: 3043–3055. [DOI] [PubMed] [Google Scholar]
- Moret F, Christiaen L, Deyts C, Blin M, Vernier P, Joly JS. 2005b. Regulatory gene expressions in the ascidian ventral sensory vesicle: evolutionary relationships with the vertebrate hypothalamus. Dev Biol 277: 567–579. [DOI] [PubMed] [Google Scholar]
- Nakane Y, Ikegami K, Iigo M, Ono H, Takeda K, Takahashi D, Uesaka M, Kimijima M, Hashimoto R, Arai N, et al. 2013. The saccus vasculosus of fish is a sensor of seasonal changes in day length. Nat Commun 4: 2108. [DOI] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. 2007. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development 134: 1151–1160. [DOI] [PubMed] [Google Scholar]
- Parmar M. 2018. Towards stem cell based therapies for Parkinson's disease. Development 145: dev156117. [DOI] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N. 2018. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360: eaaq1723. [DOI] [PubMed] [Google Scholar]
- Razy-Krajka F, Brown ER, Horie T, Callebert J, Sasakura Y, Joly JS, Kusakabe TG, Vernier P. 2012. Monoaminergic modulation of photoreception in ascidian: evidence for a proto-hypothalamo–retinal territory. BMC Biol 10: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Lu Z, Meinertzhagen IA. 2016. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. Elife 5: e16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y. 2007. Germline transgenesis and insertional mutagenesis in the ascidian Ciona intestinalis. Dev Dyn 236: 1758–1767. [DOI] [PubMed] [Google Scholar]
- Sasakura Y, Konno A, Mizuno K, Satoh N, Inaba K. 2008. Enhancer detection in the ascidian Ciona intestinalis with transposase-expressing lines of Minos. Dev Dyn 237: 39–50. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Wang W, Stolfi A. 2018. Single-cell transcriptome profiling of Ciona larval brain. bioRxiv. 10.1101/319327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. 2018. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360: 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.