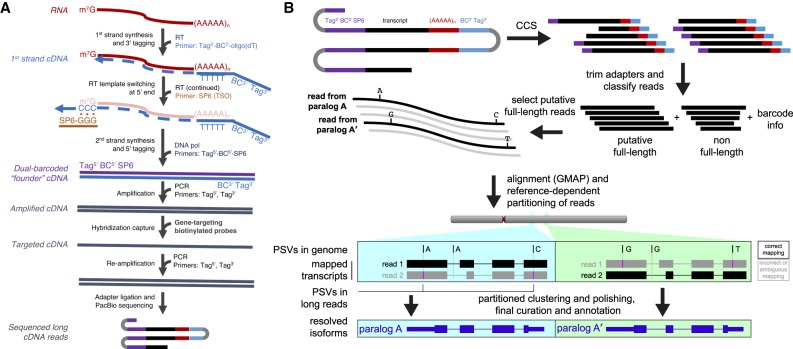
Figure 2.
Transcript capture and long-read sequencing for resolution of nearly identical duplicate genes. (A) Poly(A)+ RNA is converted to first-strand cDNA by reverse transcriptase (RT) using a specialized oligo(dT) primer containing the 3′ barcode (BC) and an outer sequence tag for later amplification. Template-independent cDNA synthesis extends the 3′ end of the cDNA with oligo-dC. RT extends the cDNA by pairing to a template switch oligo (TSO; SP6 sequence) with 3′ rG bases. Second-strand synthesis is carried out with DNA polymerase and a primer directed toward the SP6 sequence, containing the 5′ barcode and the other outer tag. After ssDNA depletion, the recovered ds-cDNA founder molecules are amplified before biotinylated probes designed to genes of interest are used for hybridization capture. A final PCR step on the target-enriched cDNA generates double-stranded molecules for long-read sequencing. (B) As part of a modified Iso-Seq workflow, sequences are first error-corrected through circular consensus sequence (CCS) generation. Then for each read, the sequences flanking the transcripts are identified and trimmed. If such sequences are present on both ends, reads are designated as putative full-length (pFL). pFL reads are mapped to the human reference (GRCh38), where the presence of multiple PSVs along the long read promotes accurate mapping even in the presence of sequencing errors. To avoid confounding paralogs, confidently mapped reads (MAPQ > 40) are partitioned into genomic segments before the Iso-Seq cluster step is performed.